6ZHA
 
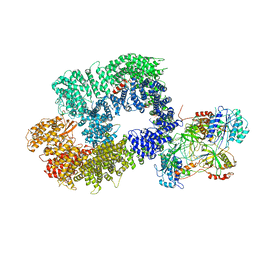 | | Cryo-EM structure of DNA-PK monomer | | Descriptor: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6HCJ
 
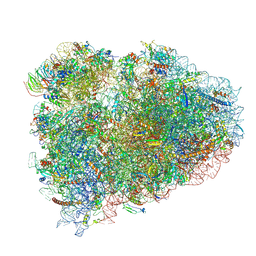 | | Structure of the rabbit 80S ribosome on globin mRNA in the rotated state with A/P and P/E tRNAs | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCQ
 
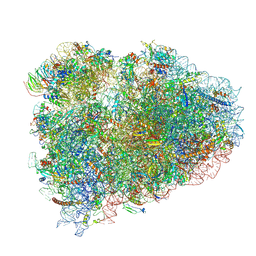 | | Structure of the rabbit collided di-ribosome (collided monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6ZHE
 
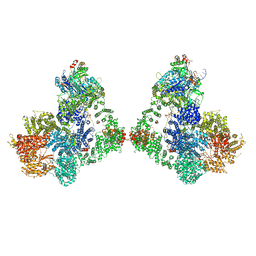 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ASV
 
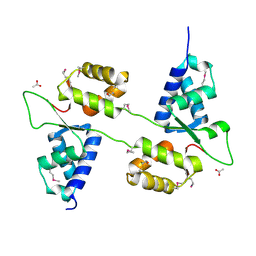 | |
7DU2
 
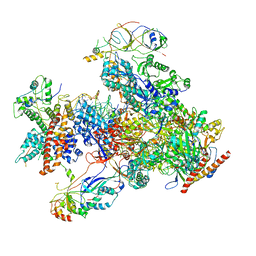 | | RNA polymerase III EC complex in post-translocation state | | Descriptor: | DNA (5'-D(P*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*T)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
1A31
 
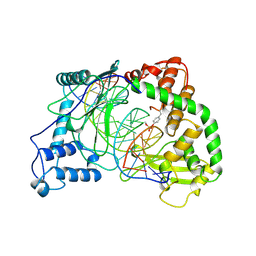 | | HUMAN RECONSTITUTED DNA TOPOISOMERASE I IN COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*5IUP*5IU*TP*GP*AP*AP*AP*AP*AP*5IUP*5IUP*5IUP*5IUP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*5IUP*5IUP*5IUP*5IUP*CP*AP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), PROTEIN (TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-27 | | Release date: | 1998-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1A49
 
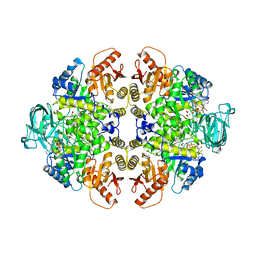 | | BIS MG-ATP-K-OXALATE COMPLEX OF PYRUVATE KINASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Larsen, T.M, Benning, M.M, Rayment, I, Reed, G.H. | | Deposit date: | 1998-02-12 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the bis(Mg2+)-ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 A resolution: ATP binding over a barrel.
Biochemistry, 37, 1998
|
|
1A35
 
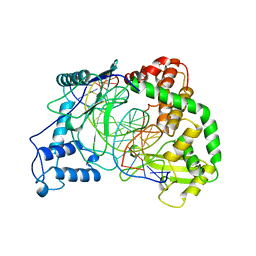 | | HUMAN TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*(BRU)P*(BRU)P*TP*TP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*+UP*+UP*+UP*+UP*CP*+UP*AP*AP*GP*TP*CP*TP*TP*TP*+ UP*T)-3'), PROTEIN (DNA TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
5GT3
 
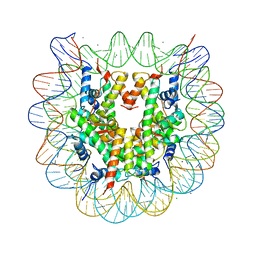 | | Crystal structure of nucleosome particle in the presence of human testis-specific histone variant, hTh2b | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-D, ... | | Authors: | Kumarevel, T, Sivaraman, P. | | Deposit date: | 2016-08-18 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural analyses of the nucleosome complexes with human testis-specific histone variants, hTh2a and hTh2b
Biophys. Chem., 221, 2017
|
|
1A36
 
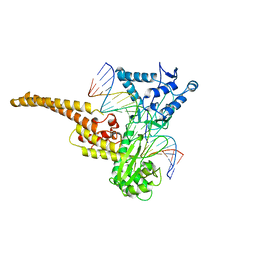 | | TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*A P*TP*TP*TP*TP*T)- 3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*AP*AP*GP*TP*C P*TP*TP*TP*TP*T)- 3'), TOPOISOMERASE I | | Authors: | Stewart, L, Redinbo, M.R, Qiu, X, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A model for the mechanism of human topoisomerase I.
Science, 279, 1998
|
|
1A5U
 
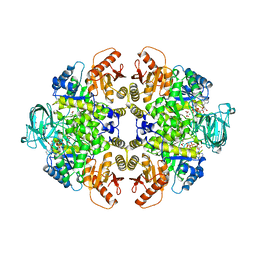 | | PYRUVATE KINASE COMPLEX WITH BIS MG-ATP-NA-OXALATE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Larsen, T.M, Benning, M.M, Rayment, I, Reed, G.H. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the bis(Mg2+)-ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 A resolution: ATP binding over a barrel.
Biochemistry, 37, 1998
|
|
1AQF
 
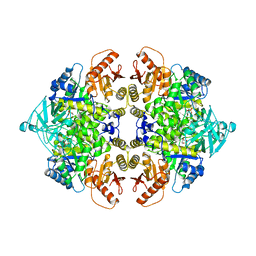 | | PYRUVATE KINASE FROM RABBIT MUSCLE WITH MG, K, AND L-PHOSPHOLACTATE | | Descriptor: | L-PHOSPHOLACTATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Larsen, T.M, Benning, M.M, Wesenberg, G.E, Rayment, I, Reed, G.H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand-induced domain movement in pyruvate kinase: structure of the enzyme from rabbit muscle with Mg2+, K+, and L-phospholactate at 2.7 A resolution.
Arch.Biochem.Biophys., 345, 1997
|
|
5FQ2
 
 | |
5IFE
 
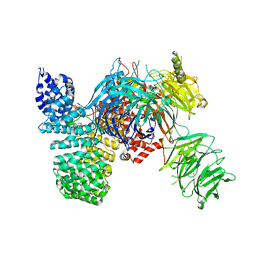 | | Crystal structure of the human SF3b core complex | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Cretu, C, Dybkov, O, De Laurentiis, E, Will, C.L, Luhrmann, R, Pena, V. | | Deposit date: | 2016-02-25 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations.
Mol.Cell, 64, 2016
|
|
2O25
 
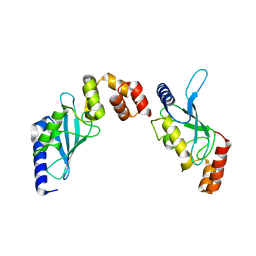 | | Ubiquitin-Conjugating Enzyme E2-25 kDa Complexed With SUMO-1-Conjugating Enzyme UBC9 | | Descriptor: | SUMO-1-conjugating enzyme UBC9, Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel and Unexpected Complex Between the SUMO-1-Conjugating Enzyme UBC9 and the Ubiquitin-Conjugating Enzyme E2-25 kDa
To be Published
|
|
5GSU
 
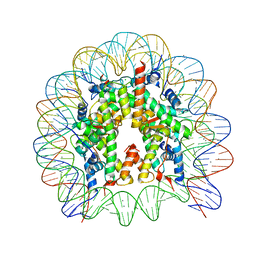 | | Crystal structure of nucleosome core particle consisting of human testis-specific histone variants, Th2A and Th2B | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-A, ... | | Authors: | Kumarevel, T, Sivaraman, P. | | Deposit date: | 2016-08-17 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analyses of the nucleosome complexes with human testis-specific histone variants, hTh2a and hTh2b
Biophys. Chem., 221, 2017
|
|
2MP2
 
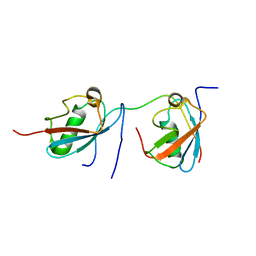 | | Solution structure of SUMO dimer in complex with SIM2-3 from RNF4 | | Descriptor: | E3 ubiquitin-protein ligase RNF4, Small ubiquitin-related modifier 3 | | Authors: | Xu, Y, Plechanovov, A, Simpson, P, Marchant, J, Leidecker, O, Sebastian, K, Hay, R.T, Matthews, S.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4.
Nat Commun, 5, 2014
|
|
2PX9
 
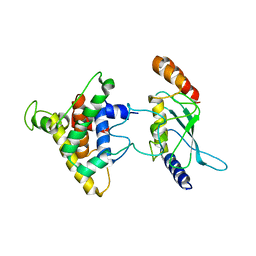 | | The intrinsic affinity between E2 and the Cys domain of E1 in Ubiquitin-like modifications | | Descriptor: | SUMO-activating enzyme subunit 2, SUMO-conjugating enzyme UBC9 | | Authors: | Wang, J.H, Hu, W.D, Cai, S, Lee, B, Song, J, Chen, Y. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications.
Mol.Cell, 27, 2007
|
|
2LYV
 
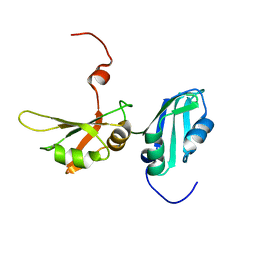 | |
6CWY
 
 | | Crystal structure of SUMO E1 in complex with an allosteric inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
1NH3
 
 | | Human Topoisomerase I Ara-C Complex | | Descriptor: | 5'-D(*(GNG)P*GP*AP*AP*AP*AP*AP*UP*UP*UP*UP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*UP*(UBB))-3', 5'-D(*AP*AP*AP*AP*AP*TP*UP*UP*UP*UP*CP*(CAR)P*AP*AP*GP*UP*CP*UP*UP*UP*UP*T)-3', ... | | Authors: | Chrencik, J.E, Burgin, A.B, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Impact of the Leukemia Drug 1-beta-D-Arabinofuranosylcytosine (Ara-C) on the Covalent Human Topoisomerase I-DNA Complex
J.Biol.Chem., 278, 2003
|
|
1LPQ
 
 | | Human DNA Topoisomerase I (70 Kda) In Non-Covalent Complex With A 22 Base Pair DNA Duplex Containing an 8-oxoG Lesion | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*(8OG)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*CP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3', DNA topoisomerase I | | Authors: | Lesher, D.T, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-05-08 | | Release date: | 2002-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | 8-Oxoguanine rearranges the active site of human topoisomerase I
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6DCL
 
 | | Crystal structure of UP1 bound to pri-miRNA-18a terminal loop | | Descriptor: | 1,2-ETHANEDIOL, Heterogeneous nuclear ribonucleoprotein A1, RNA (5'-R(*AP*GP*UP*AP*GP*AP*UP*UP*AP*GP*C)-3') | | Authors: | Kooshapur, H, Sattler, M. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1.
Nat Commun, 9, 2018
|
|
6CWZ
 
 | | Crystal structure of apo SUMO E1 | | Descriptor: | SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ZINC ION | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
