8SS2
 
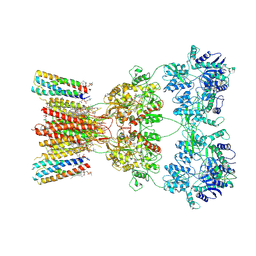 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS9
 
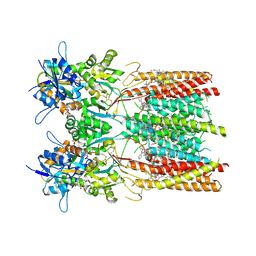 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS3
 
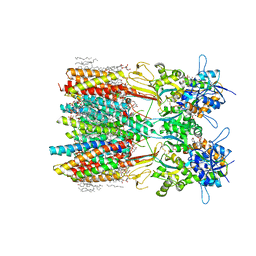 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS6
 
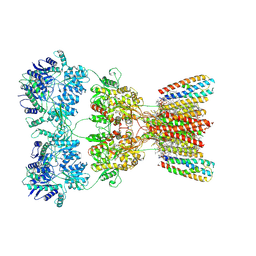 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK, channel blocker spermidine and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6U5S
 
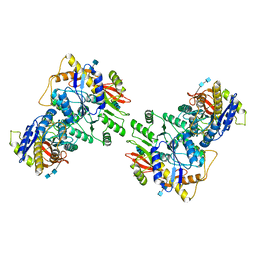 | |
6UCB
 
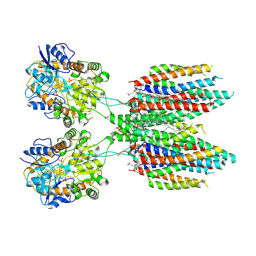 | | GluA2 in complex with its auxiliary subunit CNIH3 - with antagonist ZK200775, LBD, TMD, CNIH3, and lipids | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Glutamate receptor 2, ... | | Authors: | Nakagawa, T. | | Deposit date: | 2019-09-15 | | Release date: | 2019-12-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structures of the AMPA receptor in complex with its auxiliary subunit cornichon.
Science, 366, 2019
|
|
6UD8
 
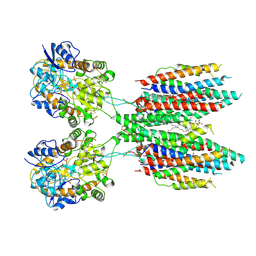 | |
6U6I
 
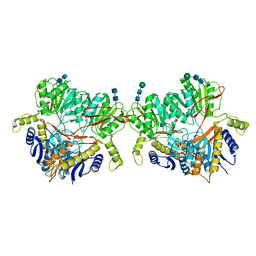 | |
6USV
 
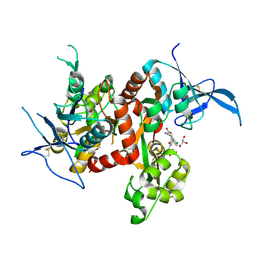 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6USU
 
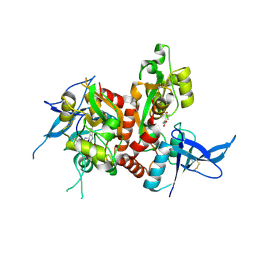 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with L689,560 and glutamate | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6UZ6
 
 | |
8USW
 
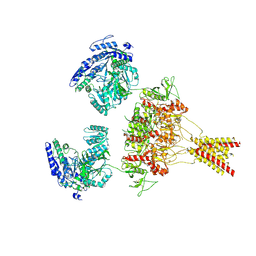 | | CNQX-bound GluN1a-3A NMDA receptor | | Descriptor: | 7-nitro-2,3-dioxo-1,2,3,4-tetrahydroquinoxaline-6-carbonitrile, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
6UZX
 
 | | Crystal structure of GLUN1/GLUN2A-4M mutant ligand-binding domain in complex with glycine and UBP791 | | Descriptor: | (2S,3R)-1-[7-(2-carboxyethyl)phenanthrene-2-carbonyl]piperazine-2,3-dicarboxylic acid, GLYCEROL, GLYCINE, ... | | Authors: | Wang, J.X, Furukawa, H. | | Deposit date: | 2019-11-15 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis of subtype-selective competitive antagonism for GluN2C/2D-containing NMDA receptors.
Nat Commun, 11, 2020
|
|
6UZG
 
 | |
8UUE
 
 | |
8USX
 
 | | Glycine-bound GluN1a-3A NMDA receptor | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 3A | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
6UD4
 
 | |
8VJ6
 
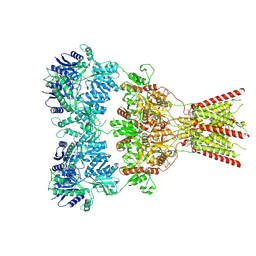 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 1 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
8VJ7
 
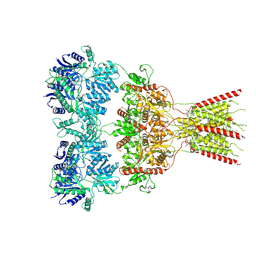 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 2 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, GLUTAMIC ACID, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.85 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
6UZR
 
 | |
6VEA
 
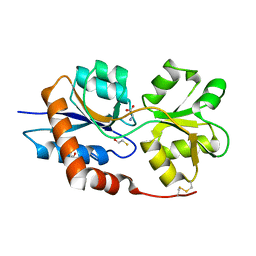 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Glycine | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCINE, Glutamate receptor 3.2, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6VE8
 
 | | Structure of the Glutamate-Like Receptor GLR3.2 ligand-binding domain in complex with Methionine | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Yoder, J.B, Sobolevsky, A.I. | | Deposit date: | 2019-12-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Arabidopsis Glutamate Receptor-like Channel GLR3.2 Ligand-Binding Domain.
Structure, 29, 2021
|
|
6UZW
 
 | | Crystal structure of GLUN1/GLUN2A ligand-binding domain in complex with glycine and UBP791 | | Descriptor: | (2S,3R)-1-[7-(2-carboxyethyl)phenanthrene-2-carbonyl]piperazine-2,3-dicarboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Wang, J.X, Furukawa, H. | | Deposit date: | 2019-11-15 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of subtype-selective competitive antagonism for GluN2C/2D-containing NMDA receptors.
Nat Commun, 11, 2020
|
|
6WHS
 
 | | GluN1b-GluN2B NMDA receptor in non-active 1 conformation at 3.95 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6WHV
 
 | | GluN1b-GluN2B NMDA receptor in complex with SDZ 220-040 and L689,560, class 2 | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
