4HC4
 
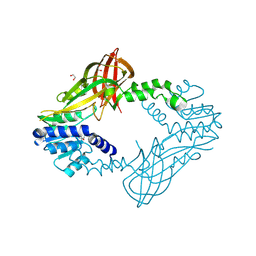 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
4H2H
 
 | | Crystal structure of an enolase (mandalate racemase subgroup, target EFI-502101) from Pelagibaca bermudensis htcc2601, with bound mg and l-4-hydroxyproline betaine (betonicine) | | Descriptor: | (2S,4R)-4-hydroxy-1,1-dimethylpyrrolidinium-2-carboxylate, (4S)-2-METHYL-2,4-PENTANEDIOL, IODIDE ION, ... | | Authors: | Vetting, M.W, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
2PLQ
 
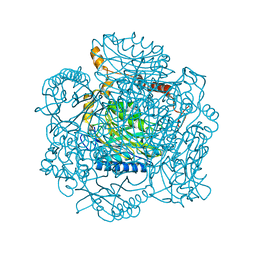 | | Crystal structure of the amidase from geobacillus pallidus RAPc8 | | Descriptor: | Aliphatic amidase | | Authors: | Kimani, S.W, Sewell, B.T, Agarkar, V.B, Sayed, M.F, Cowan, D.A. | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The quaternary structure of the amidase from Geobacillus pallidus RAPc8 is
revealed by its crystal packing.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5Z0A
 
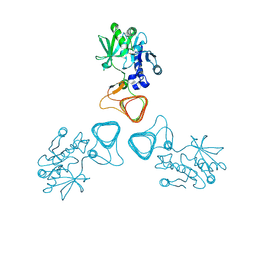 | | ST0452(Y97N)-GlcNAc binding form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dual sugar-1-phosphate nucleotidylyltransferase | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
3NRC
 
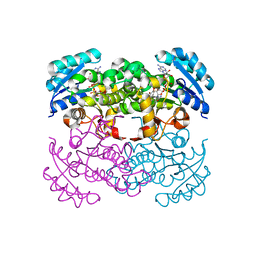 | | Crystal Structure of the Francisella tularensis enoyl-acyl carrier protein reductase (FabI) in complex with NAD+ and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase (NADH), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Mehboob, S, Santarsiero, B.D, Truong, K, Johnson, M.E. | | Deposit date: | 2010-06-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of the Francisella tularensis enoyl-acyl carrier protein reductase (FabI) in complex with NAD(+) and triclosan.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3NSG
 
 | | Crystal Structure of OmpF, an Outer Membrane Protein from Salmonella typhi | | Descriptor: | CITRATE ANION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Balasubramaniam, D, Arockiasamy, A, Sharma, A, Krishnaswamy, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Asymmetric pore occupancy in crystal structure of OmpF porin from Salmonella typhi
J.Struct.Biol., 178, 2012
|
|
9CQG
 
 | |
9CQH
 
 | |
9CQI
 
 | |
9CQJ
 
 | |
9CQF
 
 | |
7UI4
 
 | |
4IWN
 
 | | Crystal structure of a putative methyltransferase CmoA in complex with a novel SAM derivative | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, tRNA (cmo5U34)-methyltransferase | | Authors: | Aller, P, Lobley, C.M, Byrne, R.T, Antson, A.A, Waterman, D.G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3PXP
 
 | |
8SBH
 
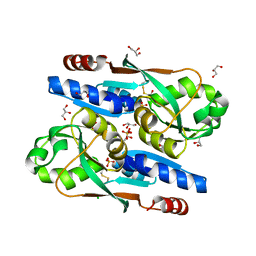 | | YeiE effector binding domain from E. coli | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Momany, C, Nune, M, Brondani, J.C, Afful, D, Neidle, E. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | FinR, a LysR-type transcriptional regulator involved in sulfur homeostasis with homologs in diverse microorganisms
To Be Published
|
|
5GVY
 
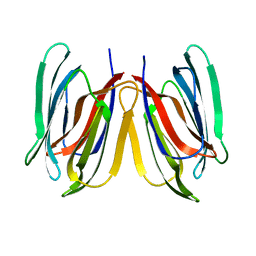 | | Crystal structure of SALT protein from Oryza sativa | | Descriptor: | Salt stress-induced protein, alpha-D-mannopyranose | | Authors: | Sharma, P, Sagar, A, Kaur, N, Sharma, I, Kirat, K, Ashish, F.N.U, Pati, P.K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | Structural insights into rice SalTol QTL located SALT protein.
Sci Rep, 10, 2020
|
|
7EZC
 
 | | Adenosine A2a receptor mutant-I92N | | Descriptor: | 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Adenosine receptor A2a,Soluble cytochrome b562 | | Authors: | Cui, M, Zhou, Q, Yao, D, Zhao, S, Song, G. | | Deposit date: | 2021-06-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of a constitutive active mutant of adenosine A 2A receptor.
Iucrj, 9, 2022
|
|
7PU1
 
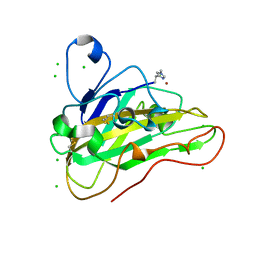 | | High resolution X-ray structure of Thermoascus aurantiacus LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Banerjee, S, Frandsen, K.E.H, Singh, R.K, Bjerrum, M.J, Lo Leggio, L. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Protonation State of an Important Histidine from High Resolution Structures of Lytic Polysaccharide Monooxygenases.
Biomolecules, 12, 2022
|
|
5JPJ
 
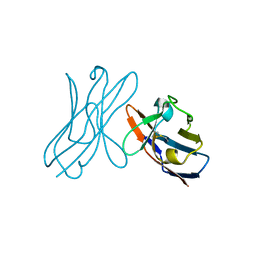 | | Crystal structure of 6aJL2-R24G | | Descriptor: | 6aJL2 protein | | Authors: | Pelaez-Aguilar, A.E, Diaz-Vilchis, A, Amero, C, Becerril, B, Rudino-Pinera, E. | | Deposit date: | 2016-05-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 6aJL2-R24G light chain variable domain: Does crystal packing explain amyloid fibril formation?
Biochem Biophys Rep, 20, 2019
|
|
3EV3
 
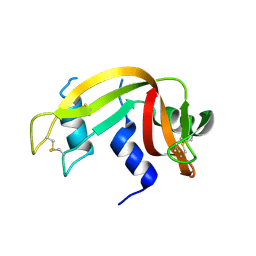 | | Crystal Structure of Ribonuclease A in 70% t-Butanol | | Descriptor: | Ribonuclease pancreatic, TERTIARY-BUTYL ALCOHOL | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
7DFQ
 
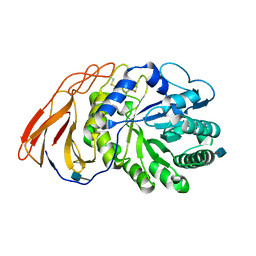 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7D1D
 
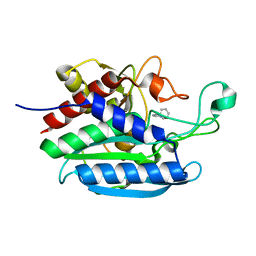 | | Crystal structure of Bacteroides thetaiotaomicron glutaminyl cyclase bound to 1-benzylimidazole | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, Glutamine cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2B
 
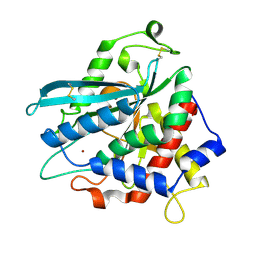 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Ni ion bound to the active site | | Descriptor: | Glutaminyl-peptide cyclotransferase, NICKEL (II) ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2I
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Fe ion bound to the active site | | Descriptor: | FE (III) ION, Glutaminyl-peptide cyclotransferase, SULFATE ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D23
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with one K ion bound to the active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutaminyl-peptide cyclotransferase, POTASSIUM ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
