2AAP
 
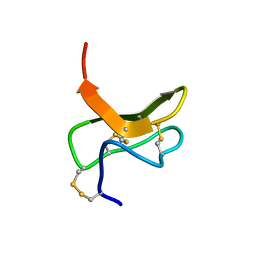 | |
1WJ1
 
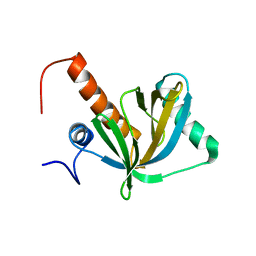 | | Solution structure of phosphotyrosine interaction domain of mouse Numb protein | | Descriptor: | Numb protein | | Authors: | Sato, M, Tomizawa, T, Koshiba, S, Tochio, N, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of phosphotyrosine interaction domain of mouse Numb protein
To be Published
|
|
1WLO
 
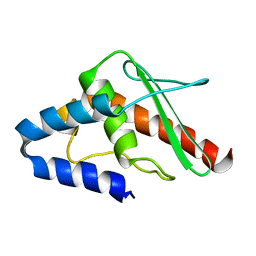 | |
2AMB
 
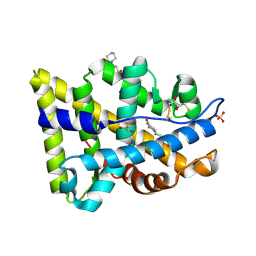 | | Crystal structure of human androgen receptor ligand binding domain in complex with tetrahydrogestrinone | | Descriptor: | 17-HYDROXY-18A-HOMO-19-NOR-17ALPHA-PREGNA-4,9,11-TRIEN-3-ONE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Pereira de Jesus-Tran, K, Cote, P.-L, Cantin, L, Blanchet, J, Labrie, F, Breton, R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity.
Protein Sci., 15, 2006
|
|
2AN2
 
 | | P332G, A333S Double mutant of the Bacillus subtilis Nitric Oxide Synthase | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, P332G A333S double mutant of Nitric Oxide Synthase from Bacillus subtilis, ... | | Authors: | Pant, K, Crane, B.R. | | Deposit date: | 2005-08-11 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a loose dimer: an intermediate in nitric oxide synthase assembly
J.Mol.Biol., 352, 2005
|
|
2ANK
 
 | | orally active thrombin inhibitors in complex with thrombin and an exosite decapeptide | | Descriptor: | N-[(1R)-2-[(1-{[({6-[AMINO(IMINO)METHYL]PYRIDIN-3-YL}METHYL)AMINO]CARBONYL}CYCLOPENTYL)AMINO]-1-(CYCLOHEXYLMETHYL)-2-OXOETHYL]GLYCINE, Thrombin heavy chain, Thrombin light chain, ... | | Authors: | Lange, U.E.W, Baucke, D, Hornberger, W, Mack, H, Seitz, W, Hoeffken, H.W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Orally active thrombin inhibitors. Part 2: optimization of the P2-moiety
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
1M42
 
 | |
2ARV
 
 | | Structure of human Activin A | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, Inhibin beta A chain, ... | | Authors: | Harrington, A.E, Morris-Triggs, S.A, Ruotolo, B.T, Robinson, C.V, Ohnuma, S, Hyvonen, M. | | Deposit date: | 2005-08-22 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of activin signalling by follistatin
Embo J., 25, 2006
|
|
1M9P
 
 | | Crystalline Human Carbonmonoxy Hemoglobin C Exhibits The R2 Quaternary State at Neutral pH In The Presence of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1WE2
 
 | | Crystal structure of shikimate kinase from mycobacterium tuberculosis in complex with MGADP and shikimic acid | | Descriptor: | 3-DEHYDROSHIKIMATE, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Pereira, J.H, de Oliveira, J.S, Canduri, F, Dias, M.V, Palma, M.S, Basso, L.A, Santos, D.S, de Azevedo Jr, W.F. | | Deposit date: | 2004-05-22 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of shikimate kinase from Mycobacterium tuberculosis reveals the binding of shikimic acid.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1M0V
 
 | | NMR STRUCTURE OF THE TYPE III SECRETORY DOMAIN OF YERSINIA YOPH COMPLEXED WITH THE SKAP-HOM PHOSPHO-PEPTIDE N-acetyl-DEpYDDPF-NH2 | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, SKAP55 homologue | | Authors: | Khandelwal, P, Keliikuli, K, Smith, C.L, Saper, M.A, Zuiderweg, E.R.P. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Biochemistry, 41, 2002
|
|
1WJM
 
 | | Solution structure of pleckstrin homology domain of human beta III spectrin. | | Descriptor: | beta-spectrin III | | Authors: | Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of pleckstrin homology domain of human beta III spectrin
To be Published
|
|
1M6E
 
 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
2AS9
 
 | | Functional and structural characterization of Spl proteases from staphylococcus aureus | | Descriptor: | ZINC ION, serine protease | | Authors: | Popowicz, G.M, Dubin, G, Stec-Niemczyk, J, Czarny, A, Dubin, A, Potempa, J, Holak, T.A. | | Deposit date: | 2005-08-23 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional and Structural Characterization of Spl Proteases from Staphylococcus aureus
J.Mol.Biol., 358, 2006
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
2ASO
 
 | | Structure of Rabbit Actin In Complex With Sphinxolide B | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WGW
 
 | | Solution Structure of the N-terminal Domain of Mouse Putative Signal Recognition Particle 54 (SRP54) | | Descriptor: | 'Signal Recognition Particle 54 | | Authors: | Li, H, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Domain of Mouse Putative Signal Recognition Particle 54 (SRP54)
To be Published
|
|
1MDY
 
 | | CRYSTAL STRUCTURE OF MYOD BHLH DOMAIN BOUND TO DNA: PERSPECTIVES ON DNA RECOGNITION AND IMPLICATIONS FOR TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*CP*AP*GP*CP*TP*GP*TP*TP*GP*A)-3'), PROTEIN (MYOD BHLH DOMAIN) | | Authors: | Ma, P.C.M, Rould, M.A, Weintraub, H, Pabo, C.O. | | Deposit date: | 1994-06-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation.
Cell(Cambridge,Mass.), 77, 1994
|
|
2AHV
 
 | | Crystal Structure of Acyl-CoA transferase from E. coli O157:H7 (YdiF)-thioester complex with CoA- 1 | | Descriptor: | COENZYME A, putative enzyme YdiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
1WMB
 
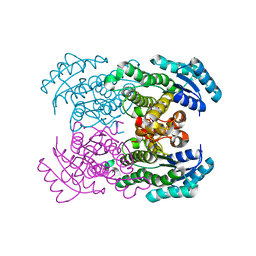 | | Crystal structure of NAD dependent D-3-hydroxybutylate dehydrogenase | | Descriptor: | CACODYLATE ION, D(-)-3-hydroxybutyrate dehydrogenase, MAGNESIUM ION | | Authors: | Ito, K, Nakajima, Y, Ichihara, E, Ogawa, K, Yoshimoto, T. | | Deposit date: | 2004-07-06 | | Release date: | 2005-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | d-3-Hydroxybutyrate Dehydrogenase from Pseudomonas fragi: Molecular Cloning of the Enzyme Gene and Crystal Structure of the Enzyme
J.Mol.Biol., 355, 2006
|
|
2AMO
 
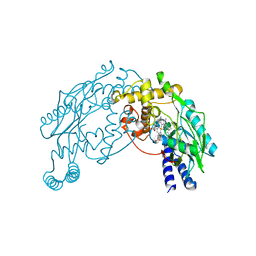 | |
2ARX
 
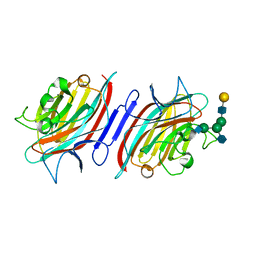 | | Pterocarpus angolensis seed lectin in complex with the decasaccharide NA2F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Buts, L, Garcia-Pino, A, Imberty, A, Amiot, N, Boons, G.-J, Lah, J, Versees, W, Wyns, L, Loris, R. | | Deposit date: | 2005-08-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of complex-type biantennary oligosaccharides by Pterocarpus angolensis lectin.
Febs J., 273, 2006
|
|
1WP9
 
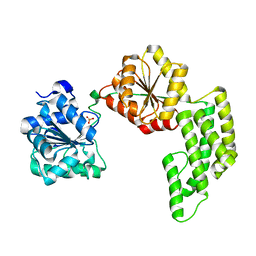 | | Crystal structure of Pyrococcus furiosus Hef helicase domain | | Descriptor: | ATP-dependent RNA helicase, putative, PHOSPHATE ION | | Authors: | Nishino, T, Komori, K, Tsuchiya, D, Ishino, Y, Morikawa, K. | | Deposit date: | 2004-08-31 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Functional Implications of Pyrococcus furiosus Hef Helicase Domain Involved in Branched DNA Processing
Structure, 13, 2005
|
|
1MK6
 
 | | SOLUTION STRUCTURE OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT MISPAIRED WITH DEOXYADENOSINE | | Descriptor: | 5'-D(*AP*CP*AP*TP*CP*GP*AP*TP*CP*T)-3', 5'-D(*AP*GP*AP*TP*AP*GP*AP*TP*GP*T)-3', 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 | | Authors: | Giri, I, Johnston, D.S, Stone, M.P. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | MISPAIRING OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT WITH DEOXYADENOSINE RESULTS IN EXTRUSION OF THE MISMATCHED DA TOWARD THE MAJOR GROOVE
Biochemistry, 41, 2002
|
|
1X4B
 
 | | Solution structure of RRM domain in Heterogeneous nuclear ribonucleaoproteins A2/B1 | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in Heterogeneous nuclear ribonucleaoproteins A2/B1
To be Published
|
|
