1UY6
 
 | | Human Hsp90-alpha with 9-Butyl-8-(3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(3,4,5-TRIMETHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1L98
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1V9W
 
 | | Solution structure of mouse putative 42-9-9 protein | | Descriptor: | putative 42-9-9 protein | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse putative 42-9-9 protein
To be Published
|
|
1L00
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1L99
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1V32
 
 | | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana | | Descriptor: | hypothetical protein RAFL09-47-K03 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-24 | | Release date: | 2004-04-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB/MDM2 domain of the hypothetical protein At5g08430 from Arabidopsis thaliana
To be Published
|
|
1V6E
 
 | | Solution Structure of a N-terminal Ubiquitin-like Domain in Mouse Tubulin-specific Chaperone B | | Descriptor: | cytoskeleton-associated protein 1 | | Authors: | Zhao, C, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a N-terminal Ubiquitin-like Domain in Mouse Tubulin-specific Chaperone B
To be Published
|
|
222L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
230L
 
 | | T4 LYSOZYME MUTANT M6L | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Gassner, N.C, Snow, S, Eldridge, A.M, Drew, D.L, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-02 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
1ZZ1
 
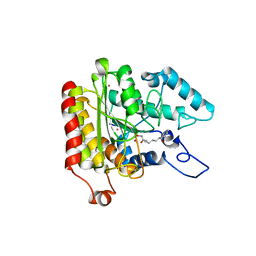 | | Crystal structure of a HDAC-like protein with SAHA bound | | Descriptor: | Histone deacetylase-like amidohydrolase, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Dickmanns, A, Schwienhorst, A, Ficner, R. | | Deposit date: | 2005-06-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of a bacterial class 2 histone deacetylase homologue
J.Mol.Biol., 354, 2005
|
|
268D
 
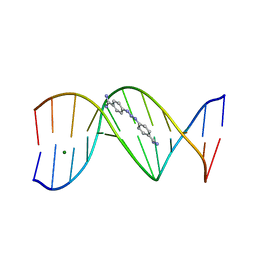 | |
1UUQ
 
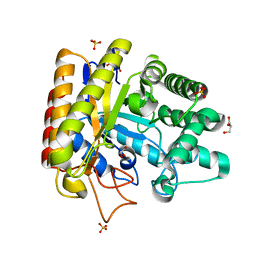 | | Exo-mannosidase from Cellvibrio mixtus | | Descriptor: | GLYCEROL, MANNOSYL-OLIGOSACCHARIDE GLUCOSIDASE, SULFATE ION | | Authors: | Dias, M.V.F, Vincent, F, Pell, G, Prates, J.A.M, Centeno, M.S.J, Ferreira, L.M.A, Gilbert, H.J, Davies, G.J, Fontes, C.M.G.A. | | Deposit date: | 2004-01-09 | | Release date: | 2004-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights Into the Molecular Determinants of Substrate Specificity in Glycoside Hydrolase Family 5 Revealed by the Crystal Structure and Kinetics of Cellvibrio Mixtus Mannosidase 5A
J.Biol.Chem., 279, 2004
|
|
227L
 
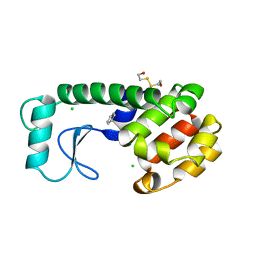 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
2A2X
 
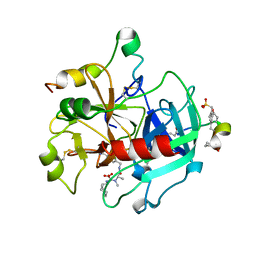 | | Orally Active Thrombin Inhibitors in Complex with Thrombin Inh12 | | Descriptor: | N-(CARBOXYMETHYL)-3-CYCLOHEXYL-D-ALANYL-N-({6-[AMINO(IMINO)METHYL]PYRIDIN-3-YL}METHYL)-N~2~-METHYL-L-ALANINAMIDE, Thrombin heavy chain, Thrombin light chain, ... | | Authors: | Lange, U.E.W, Baucke, D, Hornberger, W, Mack, H, Seitz, W, Hoeffken, H.W. | | Deposit date: | 2005-06-23 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Orally active thrombin inhibitors. Part 2: optimization of the P2-moiety
BIOORG.MED.CHEM.LETT., 16, 2006
|
|
1M33
 
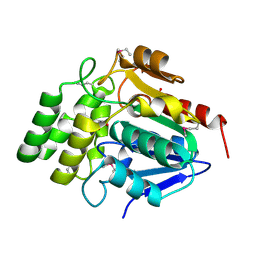 | | Crystal Structure of BioH at 1.7 A | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-PROPANOIC ACID, BioH protein | | Authors: | Sanishvili, R, Savchenko, A, Skarina, T, Edwards, A, Joachimiak, A, Yakunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-06-26 | | Release date: | 2003-01-21 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Integrating structure, bioinformatics, and enzymology to discover function: BioH, a new carboxylesterase from Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
1UWW
 
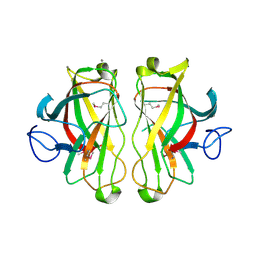 | | X-ray crystal structure of a non-crystalline cellulose specific carbohydrate-binding module: CBM28. | | Descriptor: | CALCIUM ION, ENDOGLUCANASE | | Authors: | Jamal, S, Nurizzo, D, Boraston, A, Davies, G.J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Crystal Structure of a Non-Crystalline Cellulose-Specific Carbohydrate-Binding Module: Cbm28
J.Mol.Biol., 339, 2004
|
|
1UP0
 
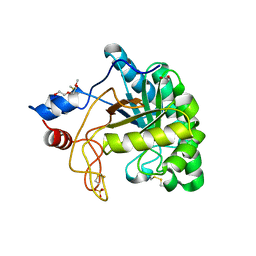 | | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with cellobiose at 1.75 angstrom | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, PUTATIVE CELLULASE CEL6, ... | | Authors: | Varrot, A, Leydier, S, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-09-26 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mycobacterium Tuberculosis Strains Possess Functional Cellulases.
J.Biol.Chem., 280, 2005
|
|
2A97
 
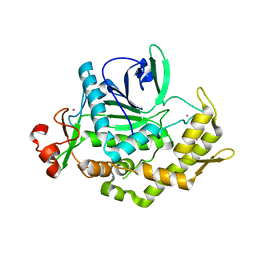 | |
1W2F
 
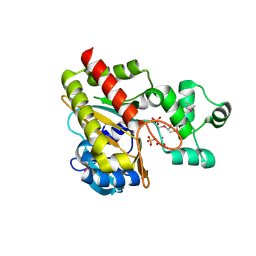 | | Human Inositol (1,4,5)-trisphosphate 3-kinase substituted with selenomethionine | | Descriptor: | INOSITOL-TRISPHOSPHATE 3-KINASE A, SULFATE ION | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
2A9X
 
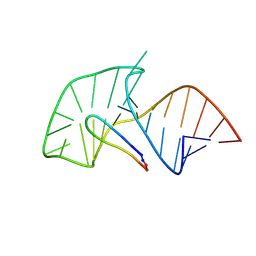 | | TAR RNA recognition by a cyclic peptidomimetic of Tat protein | | Descriptor: | BIV TAR RNA, BIV-2 cyclic peptide | | Authors: | Leeper, T.C, Athanassiou, Z, Dias, R.L, Robinson, J.A, Varani, G. | | Deposit date: | 2005-07-12 | | Release date: | 2005-11-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | TAR RNA recognition by a cyclic peptidomimetic of Tat protein.
Biochemistry, 44, 2005
|
|
1MAF
 
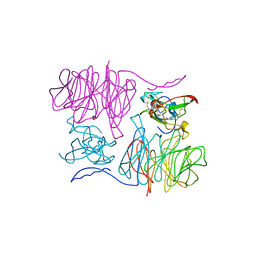 | |
1WFV
 
 | | Solution structure of the fifth PDZ domain of human membrane associated guanylate kinase inverted-2 (KIAA0705 protein) | | Descriptor: | membrane associated guanylate kinase inverted-2 | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fifth PDZ domain of human membrane associated guanylate kinase inverted-2 (KIAA0705 protein)
To be Published
|
|
2A4D
 
 | | Structure of the human ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) | | Descriptor: | Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
1WFS
 
 | | Solution Structure of Glia Maturation Factor-gamma from Mus Musculus | | Descriptor: | Glia maturation factor gamma | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
1WCK
 
 | | Crystal structure of the C-terminal domain of BclA, the major antigen of the exosporium of the Bacillus anthracis spore. | | Descriptor: | BCLA PROTEIN, CACODYLATE ION | | Authors: | Rety, S, Salamitou, S, Augusto, L.A, Chaby, R, Lehegarat, F, Lewit-Bentley, A. | | Deposit date: | 2004-11-17 | | Release date: | 2005-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The Crystal Structure of the Bacillus Anthracis Spore Surface Protein Bcla Shows Remarkable Similarity to Mammalian Proteins.
J.Biol.Chem., 280, 2005
|
|
