8AYS
 
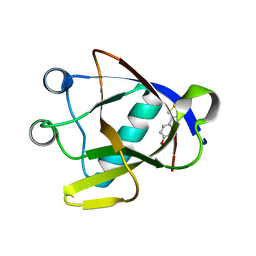 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 4-(2-aminothiazol-4-yl)phenol | | Descriptor: | 4-(2-amino-1,3-thiazol-4-yl)phenol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
8B8B
 
 | | Multimerization domain of Munia virus 1 phosphoprotein | | Descriptor: | Munia Bornavirus 1 phosphoprotein, NITRATE ION | | Authors: | Chenavier, F, Tarbouriech, N, Bourhis, J.M, Tomonaga, K, Horie, M, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
1FPT
 
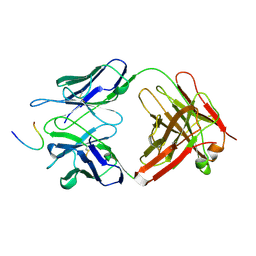 | |
1FT4
 
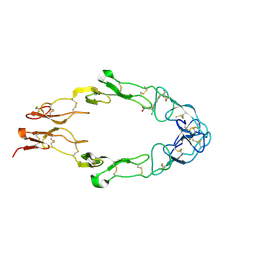 | |
6VLF
 
 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
9EB6
 
 | | Chicken YF1.7*1 presenting myristoylated peptide derived from tegument protein CIRC | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khandokar, Y, Wang, C.J.H, Rossjohn, J, Le Nours, J. | | Deposit date: | 2024-11-11 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for presentation of N-myristoylated peptides by the chicken YF1∗7.1 molecule.
J.Biol.Chem., 301, 2025
|
|
6FCK
 
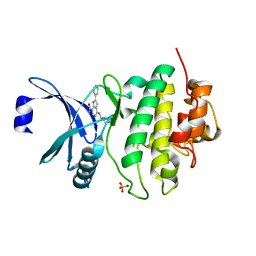 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-phenyl-4-[[(3~{S})-piperidin-3-yl]amino]-1~{H}-indole-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
3KQT
 
 | |
8W13
 
 | | Crystal structure of MYST acetyltransferase domain in complex with N-(1-(5-bromo-2-methoxyphenyl)-1H-1,2,3-triazol-4-yl)-2-methoxybenzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone acetyltransferase KAT8, ... | | Authors: | Chen, C, Dou, Y, Wang, M, Xu, C, Buesking, A. | | Deposit date: | 2024-02-15 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identification of triazolyl KAT6 inhibitors via a templated fragment approach.
Bioorg.Med.Chem.Lett., 113, 2024
|
|
1FAR
 
 | |
7ADV
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ447 (compound 6v) | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[2-(2-morpholin-4-ylethylsulfonyl)ethyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Pye, V.E, Cherepanov, P. | | Deposit date: | 2020-09-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | HIV-1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants.
Acs Infect Dis., 7, 2021
|
|
7ADU
 
 | |
7R9P
 
 | | Crystal structure of HPK1 in complex with compound 14 | | Descriptor: | 6-amino-2-fluoro-N,N-dimethyl-3-(4'-methylspiro[cyclopropane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl)benzamide, Hematopoietic progenitor kinase, SULFATE ION | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
6UY9
 
 | |
1FBA
 
 | | THE CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE FROM DROSOPHILA MELANOGASTER AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | FRUCTOSE 1,6-BISPHOSPHATE ALDOLASE | | Authors: | Piontek, K, Hester, G, Brenner-Holzach, O. | | Deposit date: | 1992-06-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of fructose-1,6-bisphosphate aldolase from Drosophila melanogaster at 2.5 A resolution.
FEBS Lett., 292, 1991
|
|
8EZX
 
 | | Lysozyme Anomalous Dataset at 293 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8TCV
 
 | | Structure of PYCR1 complexed with 4-bromobenzene-1,3-dicarboxylic acid | | Descriptor: | 4-bromobenzene-1,3-dicarboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
8TCZ
 
 | |
8TCW
 
 | |
8TCX
 
 | | Structure of PYCR1 complexed with 2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-carboxylic acid | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-carboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
8EZU
 
 | | Lysozyme Anomalous Dataset at 273 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8EZP
 
 | | Lysozyme Anomalous Dataset at 260 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8F0B
 
 | | Lysozyme Anomalous Dataset at 240 K and 7.1 keV | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Doukov, T, Yabukarski, F, Herschlag, D. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7TM5
 
 | |
8YLE
 
 | | Crystal structure of Werner syndrome helicase complexed with AMP-PCP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Yang, Y, Fu, L, Sun, X, Cheng, H, Chen, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of werner syndrome helicase complexed with AMP-PCP at 1.86 Angstroms resolution.
To Be Published
|
|
