1TOM
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, METHYL-PHE-PRO-AMINO-CYCLOHEXYLGLYCINE | | Authors: | Chen, Z. | | Deposit date: | 1996-12-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, evaluation, and crystallographic analysis of L-371,912: A potent and selective active-site thrombin inhibitor.
Bioorg.Med.Chem.Lett., 7, 1997
|
|
6PIZ
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-1 (NR02-24) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopropyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, GLYCEROL, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
3NYU
 
 | |
7ZLX
 
 | |
4D9R
 
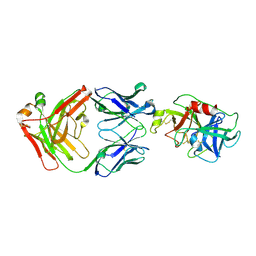 | |
6PJ2
 
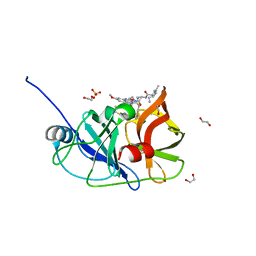 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-P5-4 (AJ-65) | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(N-acetyl-L-isoleucyl)amino]-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-N-[(1-methylcyclo propyl)sulfonyl]-5,16-dioxo-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacycl opentadecine-14a(5H)-carboxamide, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
4D8E
 
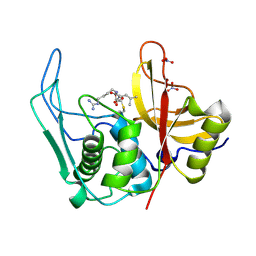 | |
4D1I
 
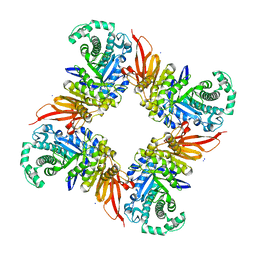 | | The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus | | Descriptor: | ACETATE ION, BETA-GALACTOSIDASE, PUTATIVE, ... | | Authors: | Larsbrink, J, Thompson, A.J, Lundqvist, M, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complex Gene Locus Enables Xyloglucan Utilization in the Model Saprophyte Cellvibrio Japonicus.
Mol.Microbiol., 94, 2014
|
|
3NYT
 
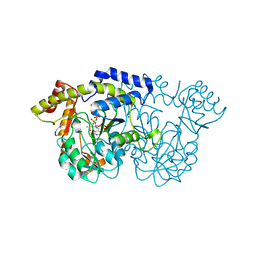 | | X-ray crystal structure of the WlbE (WpbE) aminotransferase from pseudomonas aeruginosa, mutation K185A, in complex with the PLP external aldimine adduct with UDP-3-amino-2-N-acetyl-glucuronic acid, at 1.3 angstrom resolution | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), Aminotransferase WbpE, SODIUM ION | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-07-15 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
4DEF
 
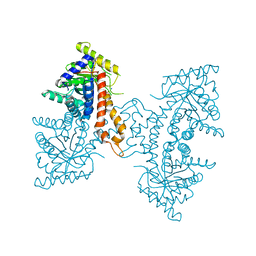 | | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from M. tuberculosis | | Descriptor: | ACETATE ION, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Capodagli, G.C, Pegan, S.D. | | Deposit date: | 2012-01-20 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Active site loop dynamics of a class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis.
Biochemistry, 52, 2013
|
|
1FMG
 
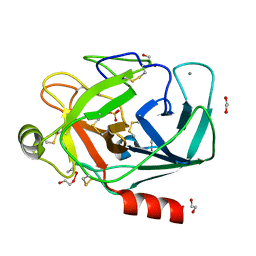 | | CRYSTAL STRUCTURE OF PORCINE BETA TRYPSIN WITH 0.04% POLYDOCANOL | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Deepthi, S, Johnson, A, Pattabhi, V. | | Deposit date: | 2000-08-17 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of porcine beta-trypsin-detergent complexes: the stabilization of proteins through hydrophilic binding of polydocanol.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FNI
 
 | | CRYSTAL STRUCTURE OF PORCINE BETA TRYPSIN WITH 0.01% POLYDOCANOL | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SULFATE ION, ... | | Authors: | Deepthi, S, Johnson, A, Pattabhi, V. | | Deposit date: | 2000-08-22 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of porcine beta-trypsin-detergent complexes: the stabilization of proteins through hydrophilic binding of polydocanol.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7ZSX
 
 | |
4DRK
 
 | | EVALUATION OF SYNTHETIC FK506 ANALOGS AS LIGANDS FOR FKBP51 AND FKBP52: COMPLEX OF FKBP51 WITH {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Gaali, S, Kress, C, Hoogeland, B, Bracher, A, Hausch, F. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Synthetic FK506 Analogues as Ligands for the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
7ZRX
 
 | |
7ZSV
 
 | |
7ZRW
 
 | |
6PIU
 
 | |
6PIY
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-2 (NR02-61) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.862 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
7ZS4
 
 | |
6PA1
 
 | | Killer cell immunoglobulin-like receptor 2DL2 in complex with HLA-C*07:02 | | Descriptor: | ARG-TYR-ARG-PRO-GLY-THR-VAL-ALA-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Moradi, S, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural plasticity of KIR2DL2 and KIR2DL3 enables altered docking geometries atop HLA-C.
Nat Commun, 12, 2021
|
|
7ZVZ
 
 | |
6PAG
 
 | | Killer cell immunoglobulin-like receptor 2DL3 in complex with HLA-C*07:02 | | Descriptor: | ARG-TYR-ARG-PRO-GLY-THR-VAL-ALA-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Moradi, S, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural plasticity of KIR2DL2 and KIR2DL3 enables altered docking geometries atop HLA-C.
Nat Commun, 12, 2021
|
|
7ZTI
 
 | |
1FNP
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> PHE FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
