8I64
 
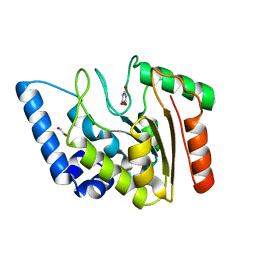 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form II | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8ZX0
 
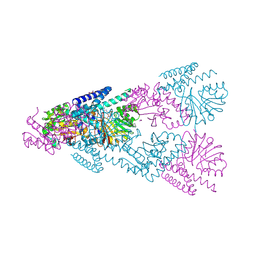 | | Crystal Structure of CntL in complex with a dual-site inhibitor | | Descriptor: | (2S)-2-[2-[[(2S,3S,4R,5R)-5-(6-azanyl-2-chloranyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]ethylamino]-3-(1H-imidazol-4-yl)propanoic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Luo, Z, Zhou, H. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structure-guided inhibitor design targeting CntL provides the first chemical validation of the staphylopine metallophore system in bacterial metal acquisition.
Eur.J.Med.Chem., 280, 2024
|
|
6WQ8
 
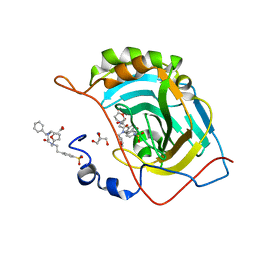 | | Carbonic Anhydrase II Complexed with 3-((2-((Furan-2-ylmethyl)(4-sulfamoylphenethyl)amino)-2-oxoethyl)(phenethyl)amino)propanoic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6X2Y
 
 | |
8B4Z
 
 | |
7RBF
 
 | | Human DNA polymerase beta crosslinked binary complex - B | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reed, A.J, Kumar, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7R2V
 
 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
7QCT
 
 | | PDZ2 of LNX2 with SARS-CoV-2_E PBM complex | | Descriptor: | Envelope small membrane protein, Ligand of Numb protein X 2 | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
6WO9
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 1-diphosphoinositol pentakisphosphate (1-IP7) and Mg | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
7C1B
 
 | | Crystal structure of a dinucleotide-binding protein (F79A/Y224A/Y246A and deletion of residues 50-75) of ABC transporter (unbound form) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sugar ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
6WO8
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5-diphosphoinositol 1,3,4,6-tetrakisphosphate (5-PP-IP4), Mg, and Fluoride ion | | Descriptor: | (1r,2R,3S,4r,5R,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G.N, Wang, H.C, Shears, S.B. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1.
Faseb J., 35, 2021
|
|
6FT2
 
 | |
6X6C
 
 | | Cryo-EM structure of NLRP1-DPP9-VbP complex | | Descriptor: | Dipeptidyl peptidase 9, NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Hollingsworth, L.R, Sharif, H, Griswold, A.R, Fontana, P, Mintseris, J, Dagbay, K.B, Paulo, J.A, Gygi, S.P, Bachovchin, D.A, Wu, H. | | Deposit date: | 2020-05-27 | | Release date: | 2021-03-10 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation.
Nature, 592, 2021
|
|
6WL5
 
 | | Crystal structure of EcmrR C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, CETYL-TRIMETHYL-AMMONIUM, CHLORIDE ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
9E6C
 
 | | Octopus sensory receptor CRT1 in complex with H3C | | Descriptor: | 1-methyl-9H-pyrido[3,4-b]indole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chemotactile receptor CRT1 | | Authors: | Jiang, H, Hibbs, R.E. | | Deposit date: | 2024-10-29 | | Release date: | 2025-07-09 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Environmental microbiomes drive chemotactile sensation in octopus.
Cell, 188, 2025
|
|
6HK8
 
 | | Crystal structure of TEX12 delta-Ctip | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Testis-expressed protein 12 | | Authors: | Salmon, L.J, Davies, O.R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-10-16 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Coiled-coil structure of meiosis protein TEX12 and conformational regulation by its C-terminal tip.
Commun Biol, 5, 2022
|
|
8ZAV
 
 | | alcohol dehydrogenases KpADH mutant - S9Y/F161K | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Ni, Y, Xu, G.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering alcohol dehydrogenases KpADH for enhanced organic-solvent tolerance and its molecular mechanisms
To Be Published
|
|
7PWC
 
 | | PARP15 catalytic domain in complex with OUL238 | | Descriptor: | 4-(cyclobutylmethoxy)benzamide, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PWA
 
 | | PARP15 catalytic domain in complex with OUL237 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(cyclopentylmethoxy)benzamide, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
8HYI
 
 | | Crystal structure of human P-cadherin MEC12 (X dimer) in complex with 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine | | Descriptor: | 2-(2-methyl-5-phenyl-1H-indole-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3, ... | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2023-01-06 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Modulation of a conformational ensemble by a small molecule that inhibits key protein-protein interactions involved in cell adhesion.
Protein Sci., 32, 2023
|
|
7PW3
 
 | | PARP15 catalytic domain in complex with OUL217 | | Descriptor: | 4-(cyclohexylmethoxy)benzamide, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Murthy, S, Lehtio, L. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
7PWK
 
 | | PARP15 catalytic domain in complex with OUL239 | | Descriptor: | 4-(cyclopropylmethoxy)benzamide, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent 2,3-dihydrophthalazine-1,4-dione derivatives as dual inhibitors for mono-ADP-ribosyltransferases PARP10 and PARP15.
Eur.J.Med.Chem., 237, 2022
|
|
6HYU
 
 | | Crystal structure of DHX8 helicase bound to single stranded poly-adenine RNA | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DHX8, RNA (5'-R(*A*AP*A)-3'), ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
8VFD
 
 | | Ternary DNA Polymerase Beta bound to DNA containing template FapydG incoming TTP analog | | Descriptor: | 1-{2-deoxy-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]-beta-D-threo-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, DNA (5'-D(*CP*CP*GP*AP*CP*(FAP)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), ... | | Authors: | Oden, P.N, Ryan, B.J, Freudenthal, B.D. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of Fapy•dG replication by Human DNA polymerase beta.
Nucleic Acids Res., 52, 2024
|
|
9FD3
 
 | | CysG(N-16)-D190N mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
