9R0X
 
 | | Crystal structure of NDM-1 with thiol compound 14a | | Descriptor: | CHLORIDE ION, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2025-04-24 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Repurposing of compound libraries yields new inhibitors of NDM-1 metallo-beta-lactamase with diverse zinc-binding moieties
Eur J Med Chem Rep, 15, 2025
|
|
4UR7
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with pyruvate | | Descriptor: | FORMIC ACID, GLYCEROL, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
9R4U
 
 | | Crystal structure of CtGH76 from Chaetomium thermophilum in complex with isomaltose | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein, ... | | Authors: | Po-Hsun, W, Essen, L.-O. | | Deposit date: | 2025-05-07 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ct GH76, a Glycoside Hydrolase 76 from Chaetomium thermophilum , with Elongated Glycan-Binding Canyon.
Int J Mol Sci, 26, 2025
|
|
9QBE
 
 | | Yeast 20S proteasome mutant: beta5_T3M in complex with ONX0914 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2025-03-02 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Proteasome-associated autoinflammatory syndromes: The impact of mutations in proteasome subunits on particle assembly, structure, and activity.
Structure, 2025
|
|
9R4N
 
 | | Crystal structure of CtGH76 from Chaetomium thermophilum | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein, ... | | Authors: | Po-Hsun, W, Essen, L.-O. | | Deposit date: | 2025-05-07 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ct GH76, a Glycoside Hydrolase 76 from Chaetomium thermophilum , with Elongated Glycan-Binding Canyon.
Int J Mol Sci, 26, 2025
|
|
9RPX
 
 | | Fragment screening of FosAKP, cryo structure in complex with fragment F2X-entry B06 | | Descriptor: | (1S)-1-(4-nitrophenyl)ethan-1-ol, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2025-06-25 | | Release date: | 2025-10-15 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
9QBY
 
 | | Yeast 20S proteasome mutant: beta5_G128V (b5-propeptide in trans) in complex with ONX0914 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2025-03-04 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Proteasome-associated autoinflammatory syndromes: The impact of mutations in proteasome subunits on particle assembly, structure, and activity.
Structure, 2025
|
|
9QDF
 
 | | Yeast 20S proteasome mutant: beta1_G128V (b1-propeptide deleted) in complex with Bortezomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2025-03-06 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Proteasome-associated autoinflammatory syndromes: The impact of mutations in proteasome subunits on particle assembly, structure, and activity.
Structure, 2025
|
|
9SKO
 
 | | Crystal structure of HLA-A0201 in complex with peptide LLWNGPMAVS | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Liu, Y, Gfeller, D, Guillaume, P, Larabi, A, Lau, K, Pojer, F. | | Deposit date: | 2025-09-02 | | Release date: | 2025-09-17 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of HLA-A0201 in complex with peptide LLWNGPMAVS
To Be Published
|
|
9R4S
 
 | | Crystal structure of CtGH76 from Chaetomium thermophilum in complex with laminaribiose | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein, ... | | Authors: | Po-Hsun, W, Essen, L.-O. | | Deposit date: | 2025-05-07 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ct GH76, a Glycoside Hydrolase 76 from Chaetomium thermophilum , with Elongated Glycan-Binding Canyon.
Int J Mol Sci, 26, 2025
|
|
9S2V
 
 | | NSP14 IN COMPLEX WITH LIGAND TDI-014925-CL-2 (compound 58) | | Descriptor: | CHLORIDE ION, Guanine-N7 methyltransferase nsp14, IMIDAZOLE, ... | | Authors: | Steinbacher, S, Huggins, D.J. | | Deposit date: | 2025-07-22 | | Release date: | 2025-09-17 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery, Optimization, and Evaluation of Non-Nucleoside SARS-CoV-2 NSP14 Inhibitors.
J.Med.Chem., 68, 2025
|
|
6UXM
 
 | |
9QWJ
 
 | | Crystal structure of S2c TCR in complex with CD1c | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karuppiah, V. | | Deposit date: | 2025-04-14 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A CD1c lipid agnostic T cell receptor bispecific engager redirects T cells against CD1c + cells.
Front Immunol, 16, 2025
|
|
9R4P
 
 | | Crystal structure of inactive (D164N) variant CtGH76 from Chaetomium thermophilum in complex with alpha-1,6-mannobiose | | Descriptor: | CtGH76, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose | | Authors: | Po-Hsun, W, Essen, L.-O. | | Deposit date: | 2025-05-07 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Ct GH76, a Glycoside Hydrolase 76 from Chaetomium thermophilum , with Elongated Glycan-Binding Canyon.
Int J Mol Sci, 26, 2025
|
|
9QF1
 
 | | Structure of CHIP E3 ubiquitin ligase TPR domain in complex with compound 2 | | Descriptor: | E3 ubiquitin-protein ligase CHIP, SULFATE ION, ~{N}-methyl-~{N}-[(1~{R})-1-pyridin-2-yl-3-pyrrolidin-1-yl-propyl]-5-[4,5,6,7-tetrakis(fluoranyl)-1~{H}-indol-3-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Breed, J. | | Deposit date: | 2025-03-11 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Discovery of Small-Molecule Ligands for the E3 Ligase STUB1/CHIP from a DNA-Encoded Library Screen.
Acs Med.Chem.Lett., 16, 2025
|
|
9QFD
 
 | | Cryo-EM structure of the fully cofilin-1-decorated actin filament (cofilactin) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2025-03-11 | | Release date: | 2025-10-08 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Choreography of rapid actin filament disassembly by coronin, cofilin, and AIP1.
Cell, 2025
|
|
7NBU
 
 | | Structure of the HigB1 toxin mutant K95A from Mycobacterium tuberculosis (Rv1955) and its target, the cspA mRNA, on the E. coli Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Giudice, E, Mansour, M, Chat, S, D'Urso, G, Gillet, R, Genevaux, P. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Substrate recognition and cryo-EM structure of the ribosome-bound TAC toxin of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
4UUQ
 
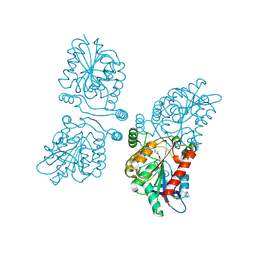 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
4URX
 
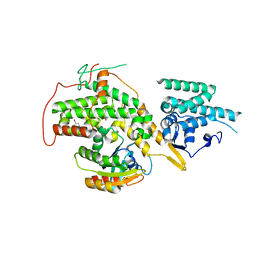 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 1-(4-bromobenzyl)pyrrolidine, 6-bromo-1H-indole, FORMIC ACID, ... | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
7RCF
 
 | |
4W51
 
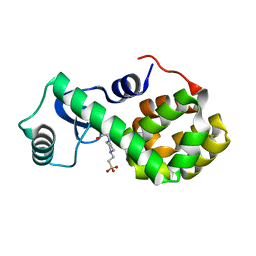 | | T4 Lysozyme L99A with No Ligand Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W53
 
 | | T4 Lysozyme L99A with Toluene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, TOLUENE | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W56
 
 | | T4 Lysozyme L99A with sec-Butylbenzene Bound | | Descriptor: | (2R)-butan-2-ylbenzene, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W59
 
 | | T4 Lysozyme L99A with n-Hexylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, hexylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W5S
 
 | | Tankyrase in complex with compound | | Descriptor: | 8-(hydroxymethyl)-2-[4-(1-methyl-1H-pyrazol-4-yl)phenyl]quinazolin-4(3H)-one, GLYCEROL, Tankyrase-1, ... | | Authors: | Johannes, J, Kazmirski, S.L, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-18 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
