4C13
 
 | | x-ray crystal structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ruane, K.M, Roper, D.I, Fulop, V, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy.
Nat.Chem.Biol., 2021
|
|
3T3U
 
 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with Bumped Kinase Inhibitor, RM-1-130 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-methoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1, ... | | Authors: | Larson, E.T, Merritt, E.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
3T4J
 
 | |
4BNH
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-hexyl- 2-phenoxyphenol | | Descriptor: | 5-hexyl-2-phenoxyphenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
4BM2
 
 | | CRYSTAL STRUCTURE OF MANGANESE PEROXIDASE 4 FROM PLEUROTUS OSTREATUS - CRYSTAL FORM II | | Descriptor: | CALCIUM ION, MANGANESE PEROXIDASE 4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2013-05-05 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Ligninolytic Peroxidase Genes in the Oyster Mushroom Genome: Heterologous Expression, Molecular Structure, Catalytic and Stability Properties, and Lignin-Degrading Ability.
Biotechnol.Biofuels, 7, 2014
|
|
3STT
 
 | |
4A30
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-BROMO-2,6-DICHLORO-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
4A6G
 
 | | N-acyl amino acid racemase from Amycalotopsis sp. Ts-1-60: G291D- F323Y mutant in complex with N-acetyl methionine | | Descriptor: | MAGNESIUM ION, N-ACETYLMETHIONINE, N-ACYLAMINO ACID RACEMASE | | Authors: | Baxter, S, Royer, S, Grogan, G, Holt-Tiffin, K.E, Taylor, I.N, Fotheringham, I.G, Campopiano, D.J. | | Deposit date: | 2011-11-02 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | An Improved Racemase/Acylase Biotransformation for the Preparation of Enantiomerically Pure Amino Acids.
J.Am.Chem.Soc., 134, 2012
|
|
4A02
 
 | | X-ray crystallographic structure of EfCBM33A | | Descriptor: | CHITIN BINDING PROTEIN | | Authors: | Vaaje-Kolstad, G, Bohle, L.A, Gaseidnes, S, Dalhus, B, Bjoras, M, Mathiesen, G, Eijsink, V.G.H. | | Deposit date: | 2011-09-07 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Characterization of the Chitinolytic Machinery of Enterococcus Faecalis V583 and High Resolution Structure of its Oxidative Cbm33 Enzyme
J.Mol.Biol., 416, 2012
|
|
4A1X
 
 | | Co-Complex structure of NS3-4A protease with the inhibitory peptide CP5-46-A (Synchrotron data) | | Descriptor: | CHLORIDE ION, CP5-46-A PEPTIDE, NONSTRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
3T3C
 
 | | Structure of HIV PR resistant patient derived mutant (comprising 22 mutations) in complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, BETA-MERCAPTOETHANOL, HIV-1 protease, ... | | Authors: | Rezacova, P, Kozisek, M, Konvalinka, J, Saskova, K.G. | | Deposit date: | 2011-07-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations in HIV-1 gag and pol Compensate for the Loss of Viral Fitness Caused by a Highly Mutated Protease.
Antimicrob.Agents Chemother., 56, 2012
|
|
4A7C
 
 | | Crystal structure of PIM1 kinase with ETP46546 | | Descriptor: | ACETATE ION, IMIDAZOLE, N-(piperidin-4-ylmethyl)-3-[3-(trifluoromethyloxy)phenyl]-[1,2,3]triazolo[4,5-b]pyridin-5-amine, ... | | Authors: | Mazzorana, M, Montoya, G. | | Deposit date: | 2011-11-12 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hit to Lead Evaluation of 1,2,3-Triazolo[4,5-B]Pyridines as Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4ADS
 
 | | Crystal structure of plasmodial PLP synthase complex | | Descriptor: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | Authors: | Guedez, G, Sinning, I, Tews, I. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4AF1
 
 | | Archeal Release Factor aRF1 | | Descriptor: | PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1, ZINC ION | | Authors: | Karlsen, J.L, Kjeldgaard, M. | | Deposit date: | 2012-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Archeal Release Factor Arf1 Contains a Metal Binding Domain
To be Published
|
|
3SQZ
 
 | |
4AL6
 
 | |
4A1G
 
 | | The crystal structure of the human Bub1 TPR domain in complex with the KI motif of Knl1 | | Descriptor: | MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1, PROTEIN CASC5 | | Authors: | Krenn, V, Wehenkel, A, Li, X, Santaguida, S, Musacchio, A. | | Deposit date: | 2011-09-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis Reveals Features of the Spindle Checkpoint Kinase Bub1-Kinetochore Subunit Knl1 Interaction.
J.Cell Biol., 196, 2012
|
|
4A1V
 
 | | Co-Complex structure of NS3-4A protease with the optimized inhibitory peptide CP5-46A-4D5E | | Descriptor: | CHLORIDE ION, CP5-46A-4D5E, NON-STRUCTURAL PROTEIN 4A, ... | | Authors: | Schmelz, S, Kuegler, J, Collins, J, Heinz, D.W. | | Deposit date: | 2011-09-20 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Affinity Peptide Inhibitors of the Hepatitis C Virus Ns3-4A Protease Refractory to Common Resistant Mutants.
J.Biol.Chem., 287, 2012
|
|
4ALI
 
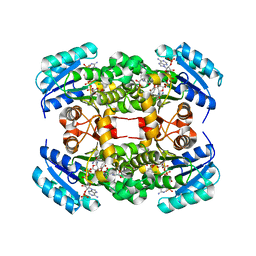 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4AL3
 
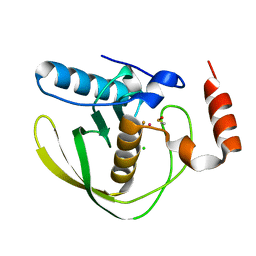 | |
4AOZ
 
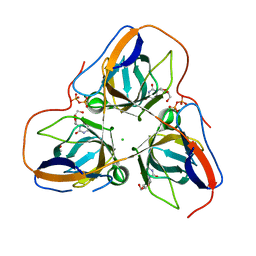 | | B. subtilis dUTPase YncF in complex with dU, PPi and Mg (P212121) | | Descriptor: | 2'-DEOXYURIDINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Timm, J, Garcia-Nafria, J, Harrison, C, Turkenburg, J.P, Wilson, K.S. | | Deposit date: | 2012-03-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tying Down the Arm in Bacillus Dutpase: Structure and Mechanism
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4A3S
 
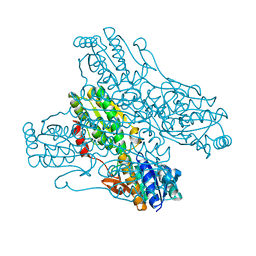 | | Crystal structure of PFK from Bacillus subtilis | | Descriptor: | 6-PHOSPHOFRUCTOKINASE | | Authors: | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | Deposit date: | 2011-10-04 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
3T37
 
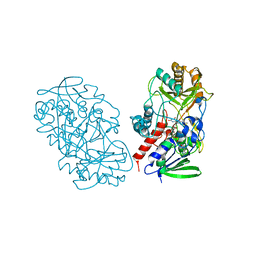 | | Crystal structure of pyridoxine 4-oxidase from Mesorbium loti | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable dehydrogenase | | Authors: | Mugo, A.N, Kobayashi, J, Mikami, B, Ohnishi, K, Yagi, T. | | Deposit date: | 2011-07-25 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structure biology and crystallization communication
TO BE PUBLISHED
|
|
3T4R
 
 | |
3T4T
 
 | |
