7X9S
 
 | | Crystal structure of a complex between the antirepressor GmaR and the transcriptional repressor MogR | | Descriptor: | GmaR, Motility gene repressor MogR | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
1U8W
 
 | | Crystal structure of Arabidopsis thaliana nucleoside diphosphate kinase 1 | | Descriptor: | Nucleoside diphosphate kinase I | | Authors: | Im, Y.J, Kim, J.-I, Shen, Y, Na, Y, Han, Y.-J, Kim, S.-H, Song, P.-S, Eom, S.H. | | Deposit date: | 2004-08-07 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Arabidopsis thaliana nucleoside diphosphate kinase-2 for phytochrome-mediated light signaling
J.Mol.Biol., 343, 2004
|
|
1MO7
 
 | | ATPase | | Descriptor: | Sodium/Potassium-transporting ATPase alpha-1 chain | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MO8
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Sodium/Potassium-Transporting ATPase alpha-1 | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1GV2
 
 | | CRYSTAL STRUCTURE OF C-MYB R2R3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-05 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
3S64
 
 | | Saposin-like protein Ac-SLP-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, Saposin-like protein 1 | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
1GV5
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-06 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1GUU
 
 | | CRYSTAL STRUCTURE OF C-MYB R1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-01-30 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
6V6T
 
 | |
3GIN
 
 | |
1GQ5
 
 | |
3GLM
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with crotonyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
1LKB
 
 | | Porcine Pancreatic Elastase/Na-Complex | | Descriptor: | CHLORIDE ION, Elastase 1, SODIUM ION, ... | | Authors: | Weiss, M.S, Panjikar, S, Nowak, E, Tucker, P.A. | | Deposit date: | 2002-04-24 | | Release date: | 2002-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding to porcine pancreatic elastase: calcium or not calcium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GQ4
 
 | |
1I0Q
 
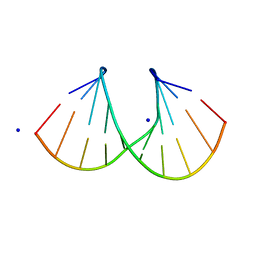 | | 1.3 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-METHYL-[TRI(OXYETHYL)] THYMINE IN PLACE OF T6, MEDIUM NA-SALT | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(126)P*AP*CP*GP*C)-3', SODIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
5X33
 
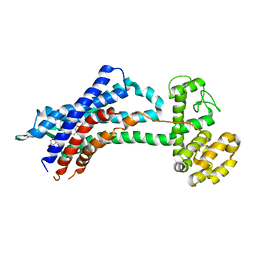 | | Leukotriene B4 receptor BLT1 in complex with BIIL260 | | Descriptor: | 4-[[3-[[4-[2-(4-hydroxyphenyl)propan-2-yl]phenoxy]methyl]phenyl]methoxy]benzenecarboximidamide, LTB4 receptor,Lysozyme,LTB4 receptor | | Authors: | Hori, T, Hirata, K, Yamashita, K, Kawano, Y, Yamamoto, M, Yokoyama, S. | | Deposit date: | 2017-02-03 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Na+-mimicking ligands stabilize the inactive state of leukotriene B4receptor BLT1.
Nat. Chem. Biol., 14, 2018
|
|
4AEH
 
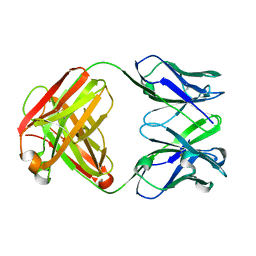 | | Crystal structure of the anti-AaHI Fab9C2 antibody | | Descriptor: | FAB ANTIBODY, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Fabrichny, I.P, Mondielli, G, Conrod, S, Martin-Eauclaire, M.F, Bourne, Y, Marchot, P. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights Into Antibody Sequestering and Neutralizing of Na+-Channel & [Alpha]-Type Modulator from Old-World Scorpion Venom
J.Biol.Chem., 287, 2012
|
|
6BUT
 
 | |
4AEI
 
 | | Crystal structure of the AaHII-Fab4C1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALPHA-MAMMAL TOXIN AAH2, CHLORIDE ION, ... | | Authors: | Fabrichny, I.P, Mondielli, G, Conrod, S, Martin-Eauclaire, M.F, Bourne, Y, Marchot, P. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into Antibody Sequestering and Neutralizing of Na+-Channel & [Alpha]-Type Modulator from Old-World Scorpion Venom
J.Biol.Chem., 287, 2012
|
|
1I0G
 
 | | 1.45 A STRUCTURE OF THE A-DECAMER GCGTATACGC WITH A SINGLE 2'-O-FLUOROETHYL THYMINE IN PLACE OF T6, MEDIUM NA-SALT | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(125)P*AP*CP*GP*C)-3', SODIUM ION | | Authors: | Tereshko, V, Wilds, C.J, Minasov, G, Prakash, T.P, Maier, M.A, Howard, A, Wawrzak, Z, Manoharan, M, Egli, M. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments.
Nucleic Acids Res., 29, 2001
|
|
1BAB
 
 | |
3GF7
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum apoprotein | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, SULFATE ION | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GF3
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaconyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GMA
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaryl-CoA | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, glutaryl-coenzyme A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
1ZUT
 
 | | Crystal Structure Of Mutant K8DP9SR58K Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
