8AKS
 
 | | 215 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKT
 
 | | 235 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AL0
 
 | | 365 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKX
 
 | | 305 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKV
 
 | | 270 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
8AKW
 
 | | 280 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural plasticity of bacterial ESCRT-III protein PspA in higher-order assemblies.
Nat.Struct.Mol.Biol., 2024
|
|
7L69
 
 | |
7LCD
 
 | |
3HB1
 
 | | Crystal structure of ed-eya2 complexed with Alf3 | | Descriptor: | ALUMINUM FLUORIDE, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
3HB0
 
 | | Structure of edeya2 complexed with bef3 | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Eyes absent homolog 2 (Drosophila), MAGNESIUM ION | | Authors: | Jung, S.K, Jeong, D.G, Ryu, S.E, Kim, S.J. | | Deposit date: | 2009-05-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor
Faseb J., 24, 2010
|
|
3U54
 
 | | Crystal structure (Type-1) of SAICAR synthetase from Pyrococcus horikoshii OT3 | | Descriptor: | 1,4-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Manjunath, K, Kanaujia, S.P, Kanagaraj, S, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2011-10-11 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of SAICAR synthetase from Pyrococcus horikoshii OT3: insights into thermal stability
Int.J.Biol.Macromol., 53, 2013
|
|
4D56
 
 | | Understanding bi-specificity of A-domains | | Descriptor: | ADENOSINE MONOPHOSPHATE, APNAA1, GLYCEROL, ... | | Authors: | Kaljunen, H, Schiefelbein, S.H.H, Stummer, D, Kozak, S, Meijers, R, Christiansen, G, Rentmeister, A. | | Deposit date: | 2014-11-03 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Elucidation of the Bispecificity of a Domains as a Basis for Activating Non-Natural Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3U55
 
 | | Crystal structure (Type-2) of SAICAR synthetase from Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, Phosphoribosylaminoimidazole-succinocarboxamide synthase, SULFATE ION | | Authors: | Manjunath, K, Kanaujia, S.P, Kanagaraj, S, Jeyakanthan, J, Sekar, K. | | Deposit date: | 2011-10-11 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SAICAR synthetase from Pyrococcus horikoshii OT3: insights into thermal stability
Int.J.Biol.Macromol., 53, 2013
|
|
6W5X
 
 | | Crystal structure of human polymerase eta complexed with N7-benzylguanine and dCTP* | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*AP*TP*(DRG)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of human polymerase eta complexed with N7-benzylguanine and dCTP*
To Be Published
|
|
6WK6
 
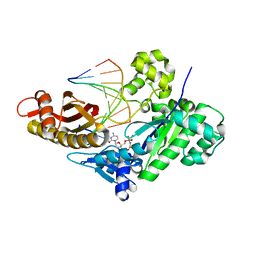 | | Crystal structure of human polymerase eta complexed with Xanthine containing DNA | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*AP*TP*(3ZO)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-04-15 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the bypass of the major deaminated purines by translesion synthesis DNA polymerase.
Biochem.J., 477, 2020
|
|
5OQT
 
 | |
4F94
 
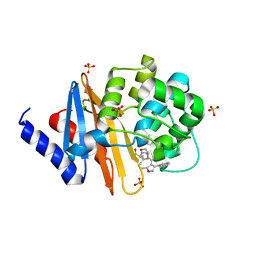 | | Structure of the Class D Beta-Lactamase OXA-24 K84D in Acyl-Enzyme Complex with Oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Beta-lactamase, SULFATE ION | | Authors: | June, C.M, Vallier, B.C, Bonomo, R.A, Leonard, D.A, Powers, R.A. | | Deposit date: | 2012-05-18 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Class D Beta-Lactamase OXA-24 K84D in Acyl-Enzyme Complex with Oxacillin
To be Published
|
|
3PH3
 
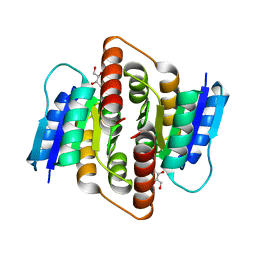 | | Clostridium thermocellum Ribose-5-Phosphate Isomerase B with d-ribose | | Descriptor: | D-ribose, Ribose-5-phosphate isomerase | | Authors: | Jung, J, Kim, J.K, Yeom, S.J, Ahn, Y.J, Oh, D.K, Kang, L.W. | | Deposit date: | 2010-11-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3PH4
 
 | | Clostridium thermocellum Ribose-5-Phosphate Isomerase B with d-allose | | Descriptor: | D-ALLOSE, Ribose-5-phosphate isomerase | | Authors: | Jung, J, Kim, J.-K, Yeom, S.-J, Ahn, Y.-J, Oh, D.-K, Kang, L.-W. | | Deposit date: | 2010-11-03 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3R6S
 
 | |
4Y46
 
 | |
6PLN
 
 | |
1S9K
 
 | |
4Y5H
 
 | |
5LW1
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_232_11_D12 in complex JNK1a1 and JIP1 peptide | | Descriptor: | ADENOSINE, C-Jun-amino-terminal kinase-interacting protein 1, DD_232_11_D12, ... | | Authors: | Wu, Y, Batyuk, A, Mittl, P.R, Honegger, A, Plueckthun, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Selective Inhibition of c-Jun N-Terminal Kinase 1 Determined by Rigid DARPin-DARPin Fusions.
J.Mol.Biol., 430, 2018
|
|
