2P9S
 
 | |
7AXU
 
 | |
2P9L
 
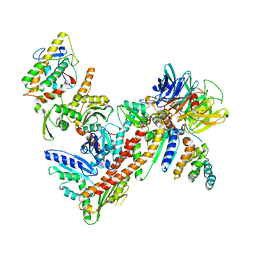 | | Crystal Structure of bovine Arp2/3 complex | | Descriptor: | Actin-like protein 2, Actin-like protein 3, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Nolen, B.J, Pollard, T.D. | | Deposit date: | 2007-03-26 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights into the Influence of Nucleotides on Actin Family Proteins from Seven Structures of Arp2/3 Complex.
Mol.Cell, 26, 2007
|
|
2OU2
 
 | | Acetyltransferase domain of Human HIV-1 Tat interacting protein, 60kDa, isoform 3 | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase HTATIP, ZINC ION | | Authors: | Min, J, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-09 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of acetyltransferase domain of Human HIV-1 Tat interacting protein in complex with acetylcoenzyme A.
To be Published
|
|
7AXQ
 
 | |
7AXP
 
 | |
7AXX
 
 | |
7AXS
 
 | |
2P02
 
 | | Crystal structure of the alpha subunit of human S-adenosylmethionine synthetase 2 | | Descriptor: | CHLORIDE ION, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthetase isoform type-2 | | Authors: | Papagrigoriou, E, Shafqat, N, Rojkova, A, Niessen, F.H, Kavanagh, K.L, von Delft, F, Gorrec, F, Ugochukwu, E, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structure of the alpha subunit of human S-adenosylmethionine synthetase 2
To be Published
|
|
5II1
 
 | | Crystal Structure of the fifth bromodomain of human polybromo (PB1) in complex with 1-methylisochromeno[3,4-c]pyrazol-5(3H)-one | | Descriptor: | 1-methyl[2]benzopyrano[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Myrianthopoulos, V, Mikros, E, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-01 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
2NP8
 
 | | Structural Basis for the Inhibition of Aurora A Kinase by a Novel Class of High Affinity Disubstituted Pyrimidine Inhibitors | | Descriptor: | N-{3-[(4-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}PYRIMIDIN-2-YL)AMINO]PHENYL}CYCLOPROPANECARBOXAMIDE, SULFATE ION, Serine/threonine-protein kinase 6 | | Authors: | Tari, L.W, Hoffman, I.D, Bensen, D.C, Hunter, M.J, Nix, J, Nelson, K.J, McRee, D.E, Swanson, R.V. | | Deposit date: | 2006-10-26 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the inhibition of Aurora A kinase by a novel class of high affinity disubstituted pyrimidine inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5ISL
 
 | |
5IVB
 
 | |
2ND0
 
 | |
5IVC
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N3 (4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid) | | Descriptor: | 1,2-ETHANEDIOL, 4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.573 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
2NML
 
 | | Crystal structure of HEF2/ERH at 1.55 A resolution | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Jin, T.C, Guo, F, Serebriiskii, I.G, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-10-21 | | Release date: | 2006-10-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
Proteins, 68, 2007
|
|
5IWF
 
 | |
5IVF
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | Descriptor: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVV
 
 | |
5IWG
 
 | | HDAC2 WITH LIGAND BRD4884 | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
2PB7
 
 | |
7EHW
 
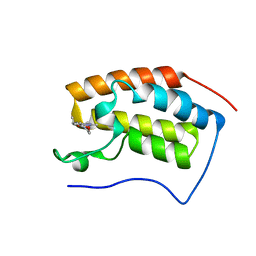 | | BRD4-BD1 in complex with LT-642-602 | | Descriptor: | 1-[4-ethyl-2-methyl-5-(6-morpholin-4-yl-1H-benzimidazol-2-yl)-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | BRD4-BD1 in complex with LT-642-602
To Be Published
|
|
2OU7
 
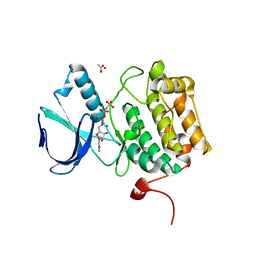 | | Structure of the Catalytic Domain of Human Polo-like Kinase 1 | | Descriptor: | ACETATE ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ding, Y.-H, Kothe, M, Kohls, D, Low, S. | | Deposit date: | 2007-02-09 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the catalytic domain of human polo-like kinase 1.
Biochemistry, 46, 2007
|
|
7EHY
 
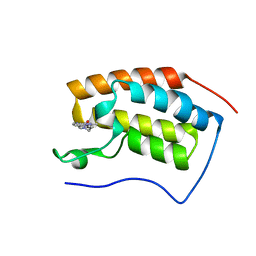 | | BRD4-BD1 in complex with LT-448-138 | | Descriptor: | 1-[2,4-dimethyl-5-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | BRD4-BD1 in complex with LT-642-602
To Be Published
|
|
7EIG
 
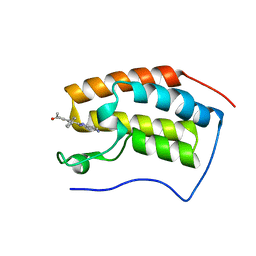 | | BRD4-BD1 in complex with LT-730-903 | | Descriptor: | 1-[5-(5-azanyl-1H-benzimidazol-2-yl)-2-methyl-4-phenyl-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | BRD4-BD1 in complex with LT-730-903
To Be Published
|
|
