5INN
 
 | | Mouse Tdp2 D358N protein, apo state with increased disorder amongst variable DNA-binding grasp conformations | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schellenberg, M.J, Appel, C.D, Williams, R.S. | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2.
Nucleic Acids Res., 44, 2016
|
|
5ODK
 
 | | Single-stranded DNA-binding protein from bacteriophage Enc34, C-terminal truncation | | Descriptor: | GLYCEROL, PHOSPHATE ION, single-stranded DNA-binding protein | | Authors: | Cernooka, E, Rumnieks, J, Kazaks, A, Tars, K. | | Deposit date: | 2017-07-05 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Basis for DNA Recognition of a Single-stranded DNA-binding Protein from Enterobacter Phage Enc34.
Sci Rep, 7, 2017
|
|
5INL
 
 | | Mouse Tdp2 reaction product (5'-phosphorylated DNA)-Mg2+ complex with deoxyadenosine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, DNA (5'-D(P*AP*CP*GP*AP*AP*TP*TP*CP*G)-3'), ... | | Authors: | Schellenberg, M.J, Vilas, C.K, Williams, R.S. | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2.
Nucleic Acids Res., 44, 2016
|
|
5INQ
 
 | |
7LAR
 
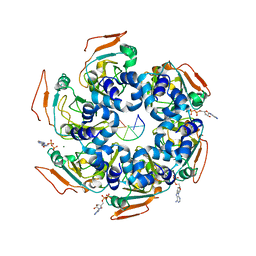 | | Cryo-EM structure of PCV2 Replicase bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent helicase Rep, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Khayat, R. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of DNA Interaction and Translocation by the Replicase of a Circular Rep-Encoding Single-Stranded DNA Virus.
Mbio, 12, 2021
|
|
7LAS
 
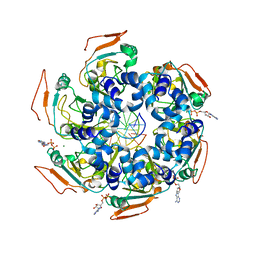 | | Cryo-EM structure of PCV2 Replicase bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent helicase Rep, DNA (5'-D(P*GP*AP*TP*CP*GP*AP*TP*CP*GP*A)-3'), ... | | Authors: | Khayat, R. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of DNA Interaction and Translocation by the Replicase of a Circular Rep-Encoding Single-Stranded DNA Virus.
Mbio, 12, 2021
|
|
6WG3
 
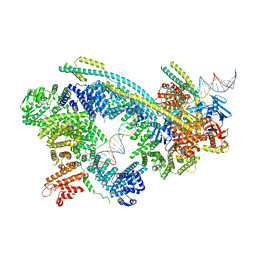 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex | | Descriptor: | Cohesin subunit SA-1, DNA (51-MER), Double-strand-break repair protein rad21 homolog, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
5INP
 
 | |
5INK
 
 | |
5INM
 
 | |
6OEQ
 
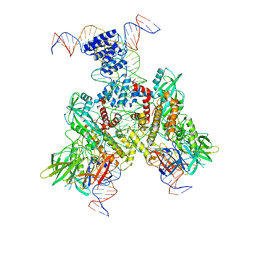 | | Cryo-EM structure of mouse RAG1/2 12RSS-PRC/23RSS-NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8UZT
 
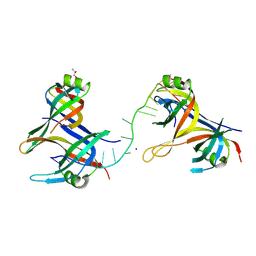 | | Mitochondrial single-stranded binding protein bound to DNA | | Descriptor: | GLYCEROL, SODIUM ION, Single-stranded DNA-binding protein, ... | | Authors: | Riccio, A.A, Pedersen, L.C, Bouvette, J, Borgnia, J.M, Copeland, W.C. | | Deposit date: | 2023-11-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the mitochondrial single-stranded DNA binding protein with DNA and DNA polymerase gamma.
Nucleic Acids Res., 52, 2024
|
|
6I1L
 
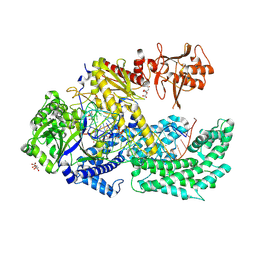 | |
6OEP
 
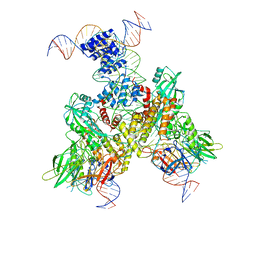 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6I1K
 
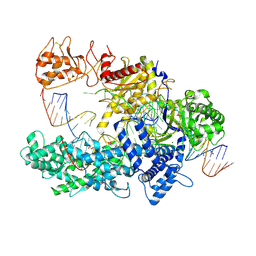 | |
6IUD
 
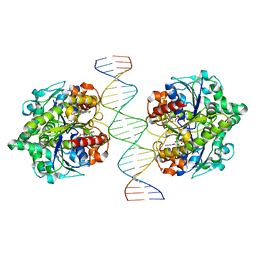 | | Structure of Helicobacter pylori Soj-ADP complex bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Yen, C.Y, Chu, C.H, Sun, Y.J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal structures of HpSoj-DNA complexes and the nucleoid-adaptor complex formation in chromosome segregation.
Nucleic Acids Res., 47, 2019
|
|
1DSA
 
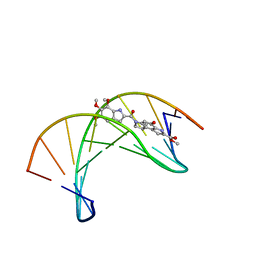 | | (+)-DUOCARMYCIN SA COVALENTLY LINKED TO DUPLEX DNA, NMR, 20 STRUCTURES | | Descriptor: | 4-HYDROXY-8-METHYL-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-3,6,7,8-TETRAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*GP*AP*CP*TP*AP*AP*TP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*AP*TP*TP*AP*GP*TP*C)-3') | | Authors: | Eis, P.S, Smith, J.A, Case, D.A, Chazin, W.J. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of a DNA duplex alkylated by the antitumor agent duocarmycin SA.
J.Mol.Biol., 272, 1997
|
|
8FYA
 
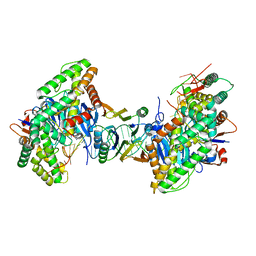 | | Cryo-EM structure of Cas1:Cas2-DEDDh:PAM-containing prespacer complex | | Descriptor: | Cas1, Cas2-DEDDh, DNA (28-MER), ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
5HLK
 
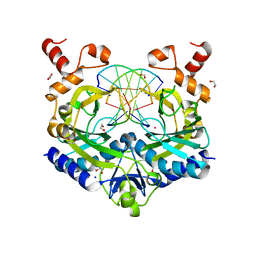 | | Crystal structure of the ternary EcoRV-DNA-Lu complex with cleaved DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*AP*GP*AP*TP)-3'), DNA (5'-D(*AP*TP*CP*TP*TP*TP)-3'), ... | | Authors: | Sangani, S.S, Kehr, A.D, Sinha, K, Rule, G.S, Jen-Jacobson, L. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Ion Binding at the Catalytic Site Induces Widely Distributed Changes in a Sequence Specific Protein-DNA Complex.
Biochemistry, 55, 2016
|
|
1DA5
 
 | |
1DA4
 
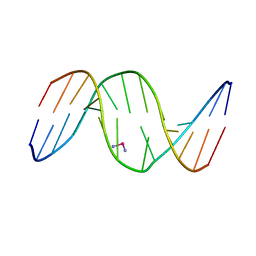 | |
6YMG
 
 | | VcaM4I restriction endonuclease in complex with 5mC-modified dsDNA | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(P*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Pastor, M, Czapinska, H, Lutz, T, Helbrecht, I, Xu, S, Bochtler, M. | | Deposit date: | 2020-04-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structures of the EVE-HNH endonuclease VcaM4I in the presence and absence of DNA.
Nucleic Acids Res., 49, 2021
|
|
6YJB
 
 | | VcaM4I restriction endonuclease 5hmC-ssDNA complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*AP*(5HC)P*AP*G)-3'), GLYCEROL, ... | | Authors: | Pastor, M, Czapinska, H, Lutz, T, Helbrecht, I, Xu, S, Bochtler, M. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the EVE-HNH endonuclease VcaM4I in the presence and absence of DNA.
Nucleic Acids Res., 49, 2021
|
|
8A71
 
 | | Crystal structure of right-handed Z-DNA containing 2'-deoxy-L-ribose in complex with the polyamine cadaverine and potassium cations at ultrahigh resolution | | Descriptor: | 5-azaniumylpentylazanium, POTASSIUM ION, Right-handed Z-DNA | | Authors: | Drozdzal, P, Manszewski, T, Gilski, M, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-06-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.69 Å) | | Cite: | Right-handed Z-DNA at ultrahigh resolution: a tale of two hands and the power of the crystallographic method.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6VBA
 
 | | Structure of human Uracil DNA Glycosylase (UDG) bound to Aurintricarboxylic acid (ATA) | | Descriptor: | 3,3'-[(3-carboxy-4-oxocyclohexa-2,5-dien-1-ylidene)methylene]bis(6-hydroxybenzoic acid), Uracil-DNA glycosylase | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An effective human uracil-DNA glycosylase inhibitor targets the open pre-catalytic active site conformation.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
