5A1K
 
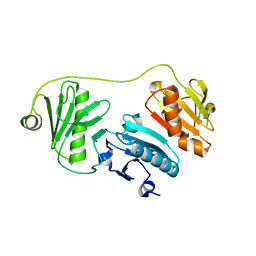 | |
2LLF
 
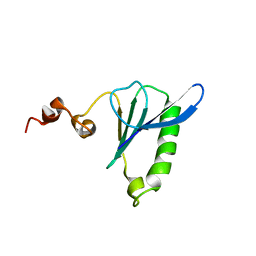 | | Sixth Gelsolin-like domain of villin in 5 mM CaCl2 | | Descriptor: | Villin-1 | | Authors: | Pfaff, D.A, Brockerman, J, Fedechkin, S, Burns, L, Zhang, F, Mcknight, C, Smirnov, S.L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Gelsolin-like activation of villin: calcium sensitivity of the long helix in domain 6.
Biochemistry, 52, 2013
|
|
2VK3
 
 | | Crystal structure of rat profilin 2a | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, PROFILIN-2, ... | | Authors: | Haikarainen, T, Kursula, P. | | Deposit date: | 2007-12-17 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, Modifications and Ligand-Binding Properties of Rat Profilin 2A.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5A1M
 
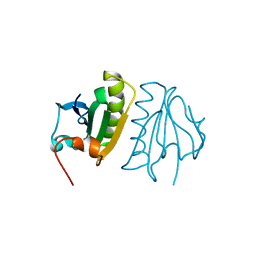 | | Crystal structure of calcium-bound human adseverin domain A3 | | Descriptor: | ADSEVERIN, CALCIUM ION | | Authors: | Chumnarnsilpa, S, Robinson, R.C, Grimes, J.M, Leyrat, C. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Calcium-Controlled Conformational Choreography in the N-Terminal Half of Adseverin.
Nat.Commun., 6, 2015
|
|
3GN4
 
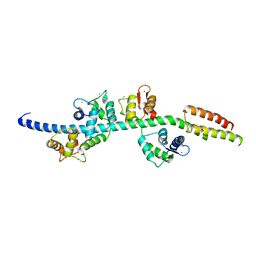 | | Myosin lever arm | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Mukherjea, M, Llinas, P, Kim, H, Travaglia, M, Safer, D, Zong, A.B, Menetrey, J, Franzini-Armstrong, C, Selvin, P.R, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach
Mol.Cell, 35, 2009
|
|
7YV9
 
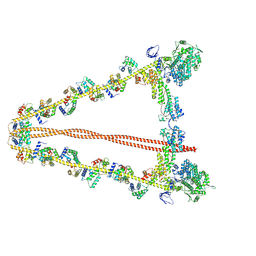 | |
3LGE
 
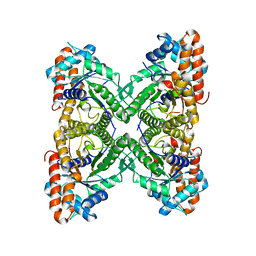 | | Crystal structure of rabbit muscle aldolase-SNX9 LC4 complex | | Descriptor: | Fructose-bisphosphate aldolase A, Sorting nexin-9 | | Authors: | Rangarajan, E.S, Park, H, Fortin, E, Sygusch, J, Izard, T. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
J.Biol.Chem., 285, 2010
|
|
2BRQ
 
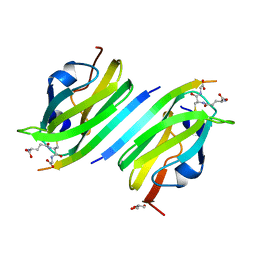 | |
3MYA
 
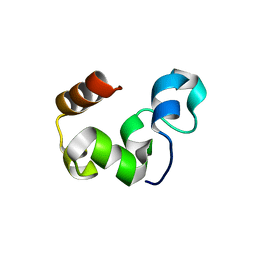 | |
3X23
 
 | | Radixin complex | | Descriptor: | Peptide from Matrix metalloproteinase-14, Radixin | | Authors: | Terawaki, S, Kitano, K, Aoyama, M, Mori, T, Hakoshima, T. | | Deposit date: | 2014-12-09 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | MT1-MMP recognition by ERM proteins and its implication in CD44 shedding
Genes Cells, 20, 2015
|
|
3MYC
 
 | |
2JF1
 
 | |
5IC0
 
 | | Structural analysis of a talin triple domain module | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Brennan, M.L. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
5IC1
 
 | | Structural analysis of a talin triple domain module, E1794Y, E1797Y, Q1801Y mutant | | Descriptor: | 1,2-ETHANEDIOL, Talin-1 | | Authors: | Wu, J, Chang, Y.-C.E, Zhang, H, Huang, Q.-Q. | | Deposit date: | 2016-02-22 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of a Talin Triple-Domain Module Suggests an Alternative Talin Autoinhibitory Configuration.
Structure, 24, 2016
|
|
7S06
 
 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosamine-1-phosphotransferase subunits alpha/beta | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2MV2
 
 | |
3I5G
 
 | | Crystal structure of rigor-like squid myosin S1 | | Descriptor: | CALCIUM ION, MALONATE ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
4MGX
 
 | |
7KFL
 
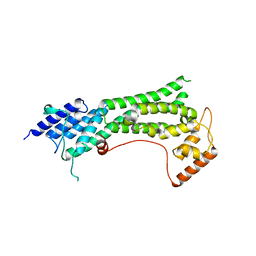 | | Crystal structure of the cargo-binding domain from the plant class XI myosin (MyoXIk) | | Descriptor: | Myosin-17 | | Authors: | Turowski, V.R, Ruiz, D.M, Nascimento, A.F.Z, Millan, C, Sammito, M.D, Juanhuix, J, Cremonesi, A.S, Uson, I, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2020-10-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the class XI myosin globular tail reveals evolutionary hallmarks for cargo recognition in plants.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3I5I
 
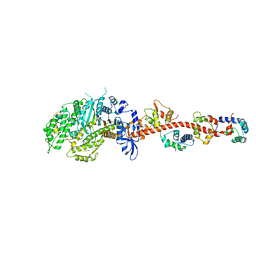 | | The crystal structure of squid myosin S1 in the presence of SO4 2- | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3I5H
 
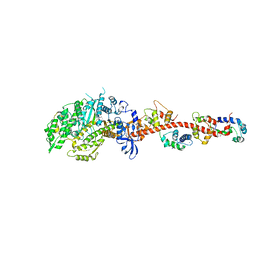 | | The crystal structure of rigor like squid myosin S1 in the absence of nucleotide | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
7YLY
 
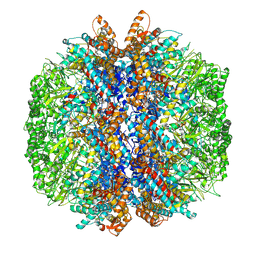 | | yeast TRiC-plp2 complex at S5 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLV
 
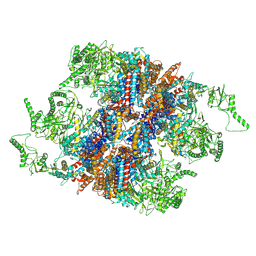 | | yeast TRiC-plp2-substrate complex at S2 ATP binding state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLU
 
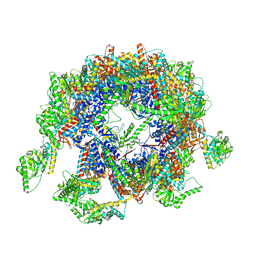 | | yeast TRiC-plp2-substrate complex at S1 TRiC-NPP state | | Descriptor: | Phosducin-like protein 2, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
7YLW
 
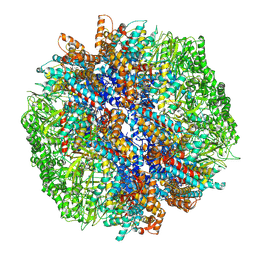 | | yeast TRiC-plp2-tubulin complex at S3 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
