4AA8
 
 | | Bovine chymosin at 1.8A resolution | | Descriptor: | CHLORIDE ION, CHYMOSIN | | Authors: | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLI
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 1 | | Descriptor: | (3-Deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-(3-deoxy-3-(3-methoxy-benzamido)-2-O-sulfo-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
2ZB0
 
 | | Crystal structure of P38 in complex with biphenyl amide inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-(3-cyanophenyl)-2'-methyl-5'-(5-methyl-1,3,4-oxadiazol-2-yl)-4-biphenylcarboxamide | | Authors: | Somers, D.O. | | Deposit date: | 2007-10-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biphenyl amide p38 kinase inhibitors 1: Discovery and binding mode
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4BTL
 
 | | Aromatic interactions in acetylcholinesterase-inhibitor complexes | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Qian, W, Engdahl, C, Berg, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4BVM
 
 | | The peripheral membrane protein P2 from human myelin at atomic resolution | | Descriptor: | CITRIC ACID, MYELIN P2 PROTEIN, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Yadav, R.P, Kursula, P. | | Deposit date: | 2013-06-26 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Atomic Resolution View Into the Structure-Function Relationships of the Human Myelin Peripheral Membrane Protein P2
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BWE
 
 | | Crystal structure of C-terminally truncated glypican-1 after controlled dehydration to 86 percent relative humidity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Glypican-1 | | Authors: | Awad, W, Svensson Birkedal, G, Thunnissen, M.M.G.M, Mani, K, Logan, D.T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Improvements in the order, isotropy and electron density of glypican-1 crystals by controlled dehydration.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
2D3B
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with AMPPNP and Methionine sulfoximine | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
2D3C
 
 | | Crystal Structure of the Maize Glutamine Synthetase complexed with ADP and Phosphinothricin Phosphate | | Descriptor: | (2S)-2-AMINO-4-[METHYL(PHOSPHONOOXY)PHOSPHORYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Unno, H, Uchida, T, Sugawara, H, Kurisu, G, Sugiyama, T, Yamaya, T, Sakakibara, H, Hase, T, Kusunoki, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Atomic Structure of Plant Glutamine Synthetase: A KEY ENZYME FOR PLANT PRODUCTIVITY
J.Biol.Chem., 281, 2006
|
|
3NVA
 
 | | Dimeric form of CTP synthase from Sulfolobus solfataricus | | Descriptor: | CTP synthase | | Authors: | Harris, P, Willemoes, M, Lauritsen, I, Johansson, E, Jensen, K.F. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structure of the dimeric form of CTP synthase from Sulfolobus solfataricus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3O0Q
 
 | | Thermotoga maritima Ribonucleotide Reductase, NrdJ, in complex with dTTP, GDP and Adenosine | | Descriptor: | ADENOSINE, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Larsson, K.-M, Logan, D.T, Nordlund, P. | | Deposit date: | 2010-07-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Adenosylcobalamin Activation in AdoCbl-Dependent Ribonucleotide Reductases.
Acs Chem.Biol., 5, 2010
|
|
1NXI
 
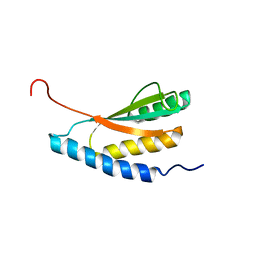 | | Solution structure of Vibrio cholerae protein VC0424 | | Descriptor: | conserved hypothetical protein VC0424 | | Authors: | Ramelot, T.A, Ni, S, Goldsmith-Fischman, S, Cort, J.R, Honig, B, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-10 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Vibrio cholerae protein VC0424: a variation of the ferredoxin-like fold.
Protein Sci., 12, 2003
|
|
3PD8
 
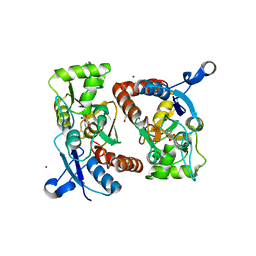 | | X-ray structure of the ligand-binding core of GluA2 in complex with (S)-7-HPCA at 2.5 A resolution | | Descriptor: | (7S)-3-hydroxy-4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridine-7-carboxylic acid, ACETIC ACID, CACODYLATE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Biostructural and pharmacological studies of bicyclic analogues of the 3-isoxazolol glutamate receptor agonist ibotenic acid.
J. Med. Chem., 53, 2010
|
|
3PZE
 
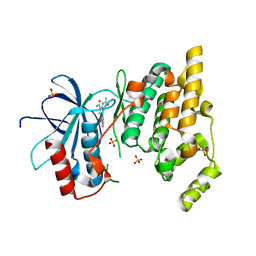 | | JNK1 in complex with inhibitor | | Descriptor: | 3-(carbamoylamino)-5-phenylthiophene-2-carboxamide, Mitogen-activated protein kinase 8, SULFATE ION | | Authors: | Xue, Y. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(piperidin-3-yl)-3-ureidothiophene-2-carboxamide (AZD7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
3PIW
 
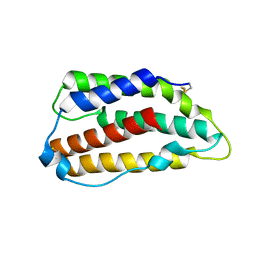 | | Zebrafish interferon 2 | | Descriptor: | Type I interferon 2 | | Authors: | Hamming, O.J, Hartmann, R, Lutfalla, G, Levraud, J.-P. | | Deposit date: | 2010-11-08 | | Release date: | 2011-07-20 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Crystal Structure of Zebrafish Interferons I and II Reveals Conservation of Type I Interferon Structure in Vertebrates.
J.Virol., 85, 2011
|
|
3OPU
 
 | |
8OZB
 
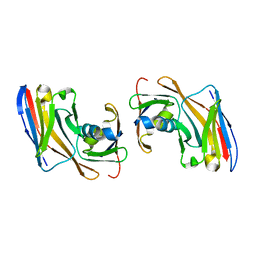 | | Crystal structure of Nup35-Nb complex | | Descriptor: | Nucleoporin NUP35, Nup35 nanobody | | Authors: | Srinivasan, V. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies.
Embo J., 43, 2024
|
|
3PL8
 
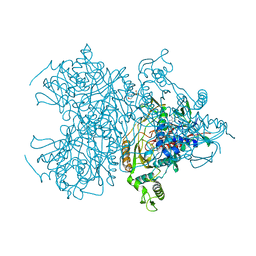 | | Pyranose 2-oxidase H167A complex with 3-deoxy-3-fluoro-beta-D-glucose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-deoxy-3-fluoro-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Divne, C. | | Deposit date: | 2010-11-14 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Regioselective control of Beta-d-glucose oxidation by pyranose 2-oxidase is intimately coupled to conformational degeneracy
J.Mol.Biol., 409, 2011
|
|
3PIV
 
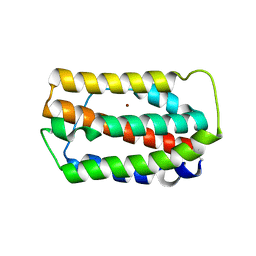 | | Zebrafish interferon 1 | | Descriptor: | Interferon, NICKEL (II) ION | | Authors: | Hamming, O.J, Hartmann, R, Lutfalla, G, Levraud, J.-P. | | Deposit date: | 2010-11-08 | | Release date: | 2011-07-20 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.086 Å) | | Cite: | Crystal Structure of Zebrafish Interferons I and II Reveals Conservation of Type I Interferon Structure in Vertebrates.
J.Virol., 85, 2011
|
|
3Q31
 
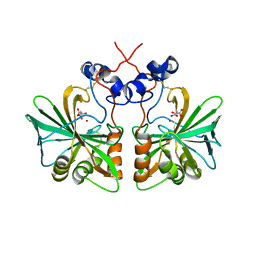 | | Structure of fungal alpha Carbonic Anhydrase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carbonic anhydrase, D-MALATE, ... | | Authors: | Cuesta-Seijo, J.A, Borchert, M.S, Navarro-Poulsen, J.C, Schnorr, K.M, Leggio, L.L. | | Deposit date: | 2010-12-21 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure of fungal alpha Carbonic Anhydrase from Aspergillus oryzae
To be Published
|
|
1Q48
 
 | | Solution NMR Structure of The Haemophilus Influenzae Iron-Sulfur Cluster Assembly Protein U (IscU) with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target IR24. This protein is not apo, it is a model without zinc binding constraints. | | Descriptor: | NifU-like protein | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shastry, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site.
J.Mol.Biol., 344, 2004
|
|
2F31
 
 | |
4L8M
 
 | | Human p38 MAP kinase in complex with a Dibenzoxepinone | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-fluoro-5-({9-[2-(morpholin-4-yl)ethoxy]-11-oxo-6,11-dihydrodibenzo[b,e]oxepin-3-yl}amino)phenyl]benzamide, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2013-06-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metabolically Stable Dibenzo[b,e]oxepin-11(6H)-ones as Highly Selective p38 MAP Kinase Inhibitors: Optimizing Anti-Cytokine Activity in Human Whole Blood.
J.Med.Chem., 56, 2013
|
|
2FQ6
 
 | |
2FSM
 
 | |
2FST
 
 | |
