6E4N
 
 | |
6E4O
 
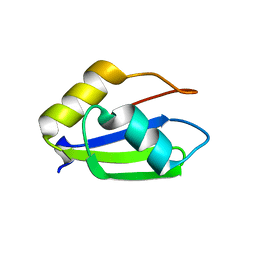 | | Structure of apo T. brucei RRM: P4(1)2(1)2 form | | Descriptor: | RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
6E4P
 
 | | Structure of the T. brucei RRM domain in complex with RNA | | Descriptor: | RNA (5'-R(P*UP*UP*UP*U)-3'), RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
6EZ7
 
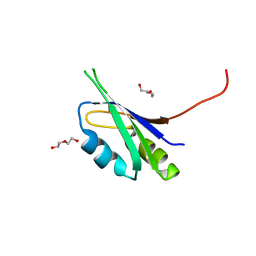 | | Pes4 RRM3 Structure | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein PES4 | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pes4 RRM3 Structure
To Be Published
|
|
6FYX
 
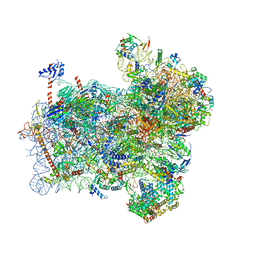 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
6FYY
 
 | | Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-03-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition.
Elife, 7, 2018
|
|
6AH0
 
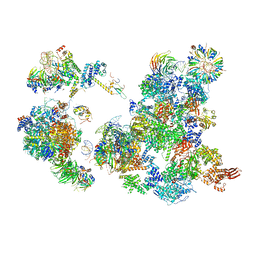 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6AHD
 
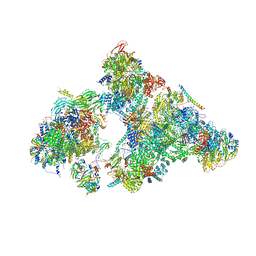 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6GD2
 
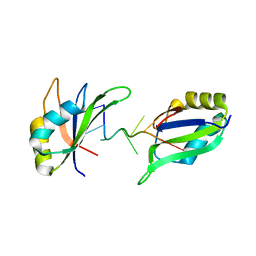 | | Structure of HuR RRM3 in complex with RNA | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*AP*UP*UP*U)-3') | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6GD3
 
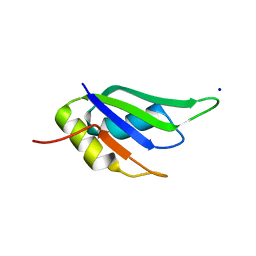 | | Structure of HuR RRM3 in complex with RNA (UAUUUA) | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*AP*UP*UP*UP*A)-3'), SODIUM ION | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6G2K
 
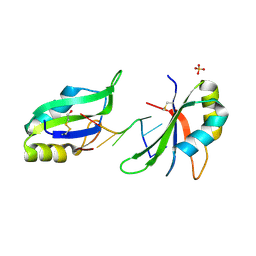 | | Structure of HuR RRM3 in complex with RNA (UUUUUU) | | Descriptor: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3'), SULFATE ION | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-03-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6GD1
 
 | | Structure of HuR RRM3 | | Descriptor: | SODIUM ION, Thioredoxin 1,ELAV-like protein 1 | | Authors: | Pabis, M, Sattler, M. | | Deposit date: | 2018-04-21 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6ELD
 
 | |
5Z57
 
 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z58
 
 | | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
6DHS
 
 | | Structure of hnRNP H qRRM1,2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Conformational Dynamics Encoded by the Inter-qRRM linker of hnRNP H.
J. Am. Chem. Soc., 2018
|
|
6DG1
 
 | |
6GML
 
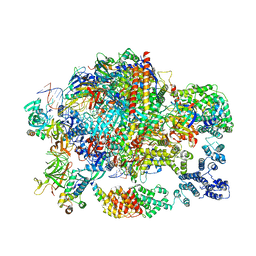 | | Structure of paused transcription complex Pol II-DSIF-NELF | | Descriptor: | DNA-directed RNA polymerase II subunit RPB9, DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Vos, S.M, Farnung, L, Urlaub, H, Cramer, P. | | Deposit date: | 2018-05-27 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of paused transcription complex Pol II-DSIF-NELF.
Nature, 560, 2018
|
|
6GX6
 
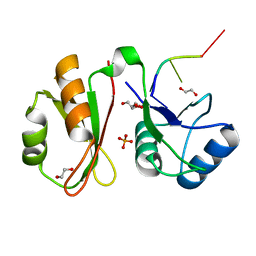 | | Crystal structure of IMP3 RRM12 in complex with RNA (ACAC) | | Descriptor: | 1,2-ETHANEDIOL, Insulin-like growth factor 2 mRNA-binding protein 3, PHOSPHATE ION, ... | | Authors: | Jia, M, Gut, H, Chao, A.J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of IMP3 RRM12 recognition of RNA.
RNA, 24, 2018
|
|
6FQ1
 
 | |
6FQR
 
 | |
5ZWO
 
 | | Cryo-EM structure of the yeast B complex at average resolution of 3.9 angstrom | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | Authors: | Bai, R, Wan, R, Yan, C, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
5ZWM
 
 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.4~4.6 angstrom (tri-snRNP and U2 snRNP Part) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
5ZWN
 
 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.3 angstrom (Part II: U1 snRNP region) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Pre-mRNA-splicing ATP-dependent RNA helicase PRP28, ... | | Authors: | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
