4XQG
 
 | |
6SM0
 
 | | Venus 66 p-Azido-L-Phenylalanin (azF) variant, dark grown | | Descriptor: | Green fluorescent protein, OXYGEN MOLECULE, SULFATE ION, ... | | Authors: | Rizkallah, P.J, Al Maslookhi, H.S, Jones, D.D. | | Deposit date: | 2019-08-21 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Stalling chromophore synthesis of the fluorescent protein Venus reveals the molecular basis of the final oxidation step.
Chem Sci, 12, 2021
|
|
6SJH
 
 | |
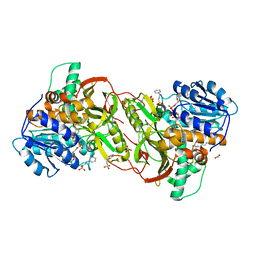
5GZU
 
 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
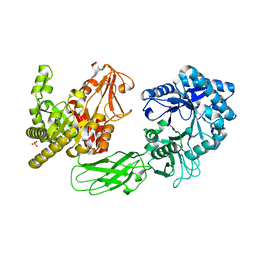
4XX0
 
 | | CoA bound to pig GTP-specific succinyl-CoA synthetase | | Descriptor: | COENZYME A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Fraser, M.E, Huang, J, Malhi, M. | | Deposit date: | 2015-01-29 | | Release date: | 2015-08-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of GTP-specific succinyl-CoA synthetase in complex with CoA.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5N1I
 
 | |
4XZF
 
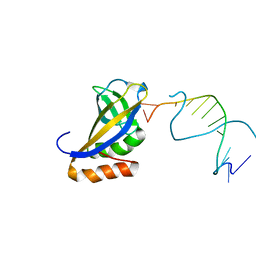 | | Crystal structure of HIRAN domain of human HLTF in complex with DNA | | Descriptor: | (DA)(DC)(DC)(DG)(DC)(DC)(DG)(DG)(DG)(DT)(DG)(DC)(DC), Helicase-like transcription factor | | Authors: | Hishiki, A, Hashimoto, H. | | Deposit date: | 2015-02-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance
J.Biol.Chem., 290, 2015
|
|
6SDW
 
 | |
4YF5
 
 | |
5N2A
 
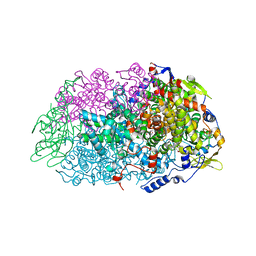 | | METHYL-COENZYME M REDUCTASE III FROM METHANOTORRIS FORMICICUS TRIGONAL FORM | | Descriptor: | 1-THIOETHANESULFONIC ACID, BROMIDE ION, Coenzyme B, ... | | Authors: | Wagner, T, Wegner, C.E, Ermler, U, Shima, S. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phylogenetic and Structural Comparisons of the Three Types of Methyl Coenzyme M Reductase from Methanococcales and Methanobacteriales.
J.Bacteriol., 199, 2017
|
|
6T28
 
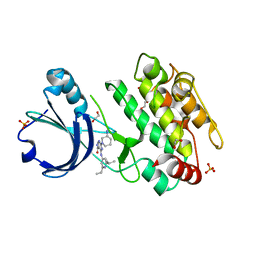 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6T29
 
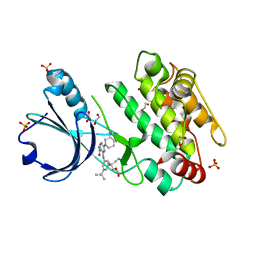 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
5NFQ
 
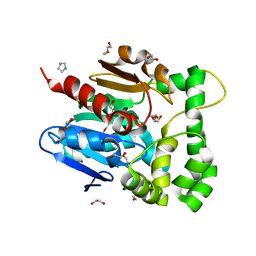 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5N4A
 
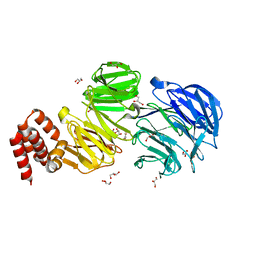 | | Crystal structure of Chlamydomonas IFT80 | | Descriptor: | GLYCEROL, Intraflagellar transport protein 80, OXALATE ION | | Authors: | Taschner, M, Mourao, A. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of intraflagellar transport protein 80 reveals a homo-dimer required for ciliogenesis.
Elife, 7, 2018
|
|
6J84
 
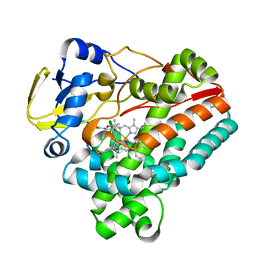 | | Crystal structure of TleB with hydroxyl analog | | Descriptor: | (2S)-2-hydroxy-N-[(2S)-1-hydroxy-3-(1H-indol-3-yl)propan-2-yl]-3-methylbutanamide, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakamura, H, Mori, T, Abe, I. | | Deposit date: | 2019-01-18 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the P450-catalyzed C-N bond formation in indolactam biosynthesis.
Nat.Chem.Biol., 15, 2019
|
|
5N5T
 
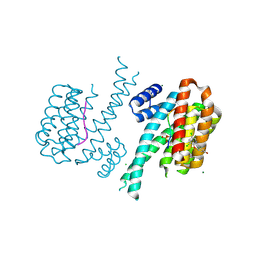 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV2 | | Descriptor: | 14-3-3 protein sigma, 4-[3,5-bis(chloranyl)pyridin-2-yl]oxyphenol, CHLORIDE ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
6J8J
 
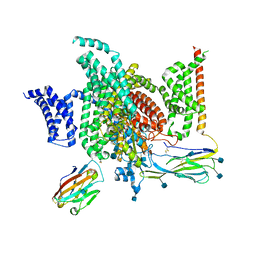 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (Y1755 down) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
5N6B
 
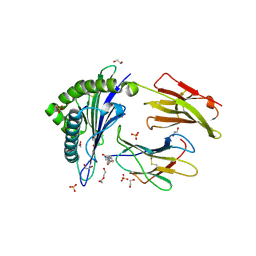 | | Human Leukocyte Antigen Class I A02 Carrying LLWNPGMAV | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Bulek, A.M, Bovay, A. | | Deposit date: | 2017-02-14 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | T cell receptor alpha variable 12-2 bias in the immunodominant response to Yellow fever virus.
Eur. J. Immunol., 48, 2018
|
|
5H2J
 
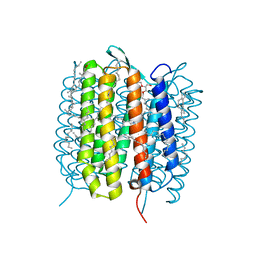 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 290 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
7AUG
 
 | | Crystal structure of rsGCamP1.3 in the ON state | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-11-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 40, 2022
|
|
5NJY
 
 | |
5GVE
 
 | | Human TOP3B-TDRD3 complex | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION, Tudor domain-containing protein 3 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.606 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
4Y9S
 
 | |
4YT4
 
 | |
5N9S
 
 | | TTK kinase domain in complex with BAY 1161909 | | Descriptor: | (2~{R})-2-(4-fluorophenyl)-~{N}-[4-[2-[(2-methoxy-4-methylsulfonyl-phenyl)amino]-[1,2,4]triazolo[1,5-a]pyridin-6-yl]phenyl]propanamide, Dual specificity protein kinase TTK | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
