7D1F
 
 | |
6MBG
 
 | |
5AKB
 
 | | MutS in complex with the N-terminal domain of MutL - crystal form 1 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.71 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AF5
 
 | | Structure of Lys33-linked triUb S.G. P 212121 | | Descriptor: | POLYUBIQUITIN-C | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
6MC4
 
 | |
7DHS
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1R)-1-phenylethyl]benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Wu, T, Xiang, Q, Wang, C, Wu, C, Zhang, C, Zhang, M, Liu, Z, Zhang, Y, Xiao, L, Xu, Y. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Y06014 is a selective BET inhibitor for the treatment of prostate cancer.
Acta Pharmacol.Sin., 42, 2021
|
|
5A3B
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, ALANINE, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
6MD4
 
 | |
7DAI
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6LS1
 
 | | Ribonuclease from Hericium erinaceus active and GMP binding form | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE, Ribonuclease T1, ... | | Authors: | Takebe, K, Suzuki, M, Sangawa, T, Kobayashi, H, Itagaki, T. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Ribonuclease from Hericium erinaceus active and GMP binding form
To Be Published
|
|
5ADD
 
 | | Structure of rat neuronal nitric oxide synthase d597n m336v mutant heme domain in complex with 7-((3-(methylamino)methyl) phenoxy)methyl)quinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[3-(methylaminomethyl)phenoxy]methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
5AB7
 
 | |
7D33
 
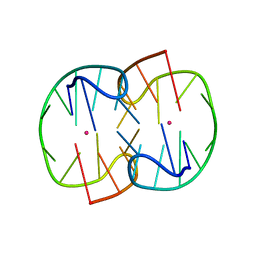 | | The Pb2+ complexed structure of TBA G8C mutant | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*CP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
5AD4
 
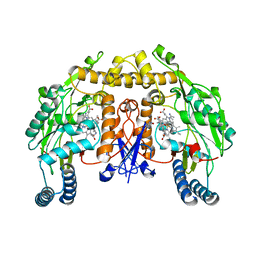 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-((3-(2-(Dimethylamino)ethyl)phenoxy)methyl)quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[3-[2-(dimethylamino)ethyl]phenoxy]methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-08-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
5AGO
 
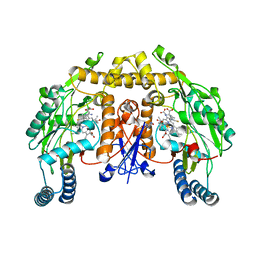 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-mercaptoacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-MERCAPTOACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
7DAL
 
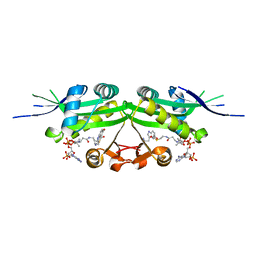 | | The crystal structure of a serotonin N-acetyltransferase in complex with serotonin and acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, SEROTONIN, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAK
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with 5-Methoxytryptamine and acetyl-CoA from Oryza Sativa | | Descriptor: | 2-(5-methoxy-1H-indol-3-yl)ethanamine, ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7D4V
 
 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase Y91F/C120S variant | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4ZVZ
 
 | | Co-crystal structures of PP5 in complex with 5-methyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid | | Descriptor: | (1R,2S,3R,4S,5S)-5-(propoxymethyl)-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Chattopadhyay, D, Swingle, M.R, Salter, E.A, Wierzbicki, A, Honkanen, R.E. | | Deposit date: | 2015-05-18 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutagenesis of PPP-family ser/thr protein phosphatases elucidate the selectivity of cantharidin and novel norcantharidin-based inhibitors of PP5C.
Biochem. Pharmacol., 109, 2016
|
|
7D64
 
 | |
4ZYP
 
 | | Crystal Structure of Motavizumab and Quaternary-Specific RSV-Neutralizing Human Antibody AM14 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | AM14 antibody Fab heavy chain, AM14 antibody light chain, Fusion glycoprotein F0,Fibritin, ... | | Authors: | Gilman, M.S.A, McLellan, J.S. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein.
Plos Pathog., 11, 2015
|
|
4ZZM
 
 | | Human ERK2 in complex with an irreversible inhibitor | | Descriptor: | 7-ethylsulfonyl-N-(oxan-4-yl)-6,8-dihydro-5H-pyrido[3,4-d]pyrimidin-2-amine, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
7KRE
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 4-((6-cyanonaphthalen-1-yl)oxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ704) | | Descriptor: | 4-[(6-cyanonaphthalen-1-yl)oxy]-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
5A6X
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with alpha-methyl-fucoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-L-fucopyranoside | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
7D75
 
 | |
