3UB3
 
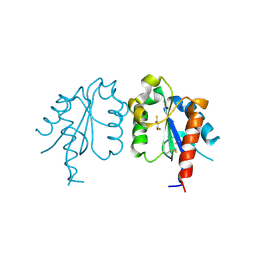 | | D96N variant of TIR domain of Mal/TIRAP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Shen, Y, Lin, Z. | | Deposit date: | 2011-10-23 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights into TIR Domain Specificity of the Bridging Adaptor Mal in TLR4 Signaling
Plos One, 7, 2012
|
|
3SKA
 
 | |
2PK8
 
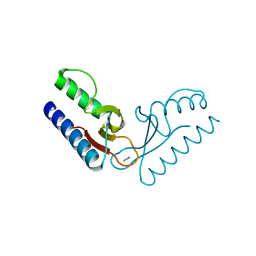 | | Crystal structure of an uncharacterized protein PF0899 from Pyrococcus furiosus | | Descriptor: | GOLD (I) CYANIDE ION, Uncharacterized protein PF0899 | | Authors: | Liu, Z.J, Tempel, W, Chen, L, Shah, A, Lee, D, Clancy-Kelley, L.L, Dillard, B.D, Rose, J.P, Sugar, F.J, Jenny Jr, F.E, Lee, H.S, Izumi, M, Shah, C, Poole III, F.L, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the hypothetical protein PF0899 from Pyrococcus furiosus at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3FZI
 
 | |
3QU8
 
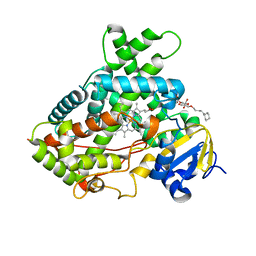 | | Crystal structure of a human cytochrome P450 2B6 (Y226H/K262R) in complex with the inhibitor 4-(4-Nitrobenzyl)pyridine. | | Descriptor: | 4-(4-nitrobenzyl)pyridine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cytochrome P450 2B6 Bound to 4-Benzylpyridine and 4-(4-Nitrobenzyl)pyridine: Insight into Inhibitor Binding and Rearrangement of Active Site Side Chains.
Mol.Pharmacol., 80, 2011
|
|
3ZIY
 
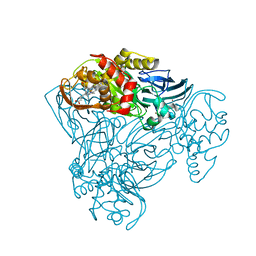 | | Structure of three-domain heme-Cu nitrite reductase from Ralstonia pickettii at 1.01 A resolution | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | Authors: | Antonyuk, S.V, Han, C, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2013-01-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
1WVF
 
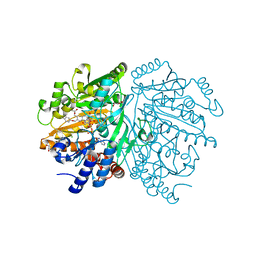 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | Descriptor: | 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
1JVB
 
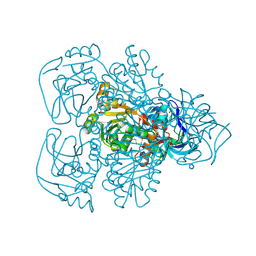 | | ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS | | Descriptor: | NAD(H)-DEPENDENT ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Esposito, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-08-29 | | Release date: | 2002-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the alcohol dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus at 1.85 A resolution.
J.Mol.Biol., 318, 2002
|
|
5F4Z
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | Descriptor: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
5W38
 
 | | 1.80A resolution structure of human IgG3 Fc (N392K) | | Descriptor: | CITRATE ANION, Immunoglobulin heavy constant gamma 3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Shah, I, Tolbert, T.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the Man5 glycoform of human IgG3 Fc.
Mol. Immunol., 92, 2017
|
|
3TXZ
 
 | | OYE1-W116Q complexed with R-carvone | | Descriptor: | (5R)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-one, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Sullivan, B, Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2011-09-23 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X‑ray Crystallography Reveals How Subtle Changes Control the Orientation of Substrate Binding in an Alkene Reductase
ACS CATALYSIS, 3, 2013
|
|
3L8P
 
 | | Crystal structure of cytoplasmic kinase domain of Tie2 complexed with inhibitor CEP11207 | | Descriptor: | 2-methyl-11-(1-methylethyl)-8-[(2S)-tetrahydro-2H-pyran-2-yl]-2,11,12,13-tetrahydro-4H-indazolo[5,4-a]pyrrolo[3,4-c]carbazol-4-one, Angiopoietin-1 receptor | | Authors: | Fedorov, A.A, Fedorov, E.V, Pauletti, D, Meyer, S.L, Hudkins, R.L, Almo, S.C. | | Deposit date: | 2010-01-03 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of cytoplasmic kinase domain of Tie2 complexed with inhibitor CEP11207
To be Published
|
|
7WQM
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 24 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(2-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
3TKH
 
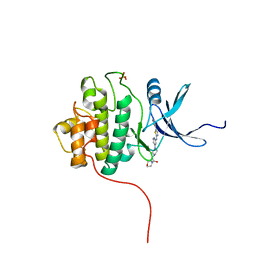 | | Crystal structure of Chk1 in complex with inhibitor S01 | | Descriptor: | 1-(morpholin-4-yl)-2-[4-(2-{[5-(pyridin-3-yl)-1,3-thiazol-2-yl]amino}pyridin-4-yl)piperazin-1-yl]ethanone, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Ikuta, M. | | Deposit date: | 2011-08-26 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Pyridyl aminothiazoles as potent inhibitors of Chk1 with slow dissociation rates.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7WQR
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 28 | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ~{N}-(3-chlorophenyl)-2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]benzamide | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-25 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
7WQS
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound 25 | | Descriptor: | 2-[(3,5-dimethyl-1,2-oxazol-4-yl)methoxy]-~{N}-(3-methoxyphenyl)benzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Sun, H, Fang, P. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of highly potent and specific AKR1C3 inhibitors to restore the chemosensitivity of drug-resistant breast cancer.
Eur.J.Med.Chem., 247, 2022
|
|
1PTJ
 
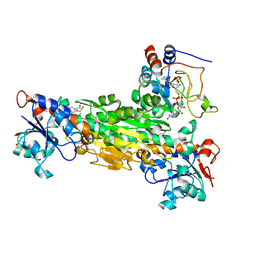 | | Crystal structure analysis of the DI and DIII complex of transhydrogenase with a thio-nicotinamide nucleotide analogue | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Singh, A, Venning, J.D, Quirk, P.G, van Boxel, G.I, Rodrigues, D.J, White, S.A, Jackson, J.B. | | Deposit date: | 2003-06-23 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Interactions between transhydrogenase and thio-nicotinamide analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation
J.Biol.Chem., 278, 2003
|
|
3P5Z
 
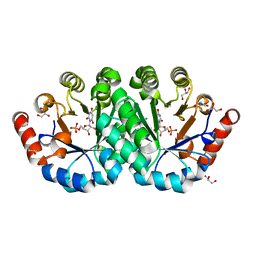 | | Crystal structure of the mutant T159S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, N-PROPANOL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-11 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3TZ6
 
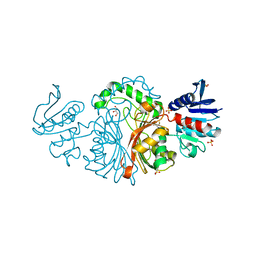 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With inhibitor SMCS (CYS) And Phosphate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CYSTEINE, GLYCEROL, ... | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OJF
 
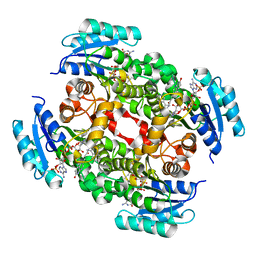 | | Crystal Structure of the Bacillus cereus Enoyl-Acyl Carrier Protein Reductase with NADP+ and indole naphthyridinone (Complex form) | | Descriptor: | (2E)-N-[(1,2-dimethyl-1H-indol-3-yl)methyl]-N-methyl-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)prop-2-enamide, Enoyl-[acyl-carrier-protein] reductase (FabL) (NADPH), NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kim, S.J, Ha, B.H, Kim, K.H, Hong, S.K, Suh, S.W, Kim, E.E. | | Deposit date: | 2010-08-22 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimeric and tetrameric forms of enoyl-acyl carrier protein reductase from Bacillus cereus
Biochem.Biophys.Res.Commun., 400, 2010
|
|
7E5Z
 
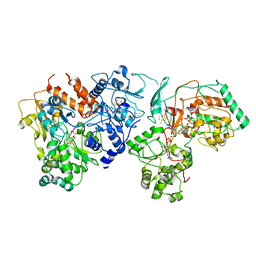 | | Dehydrogenase holoenzyme | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Roh, S.H, Park, J.S, Heo, Y.Y. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dehydrogenase holoenzyme
To Be Published
|
|
3L5M
 
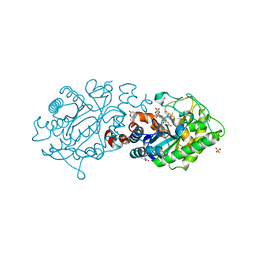 | | Xenobiotic reductase A - coumarin bound | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-22 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
1I0H
 
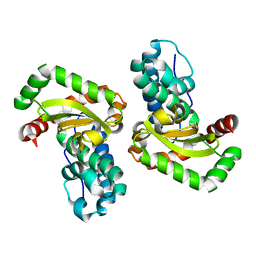 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE MUTANT Y174F AT 1.35 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE Y174F MUTANT | | Authors: | Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2001-01-29 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Removing a hydrogen bond in the dimer interface of Escherichia coli manganese superoxide dismutase alters structure and reactivity.
Biochemistry, 40, 2001
|
|
3LDW
 
 | | Crystal Structure of Plasmodium vivax geranylgeranylpyrophosphate synthase PVX_092040 with zoledronate and IPP bound | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Wernimont, A.K, Lew, J, Zhao, Y, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
3LEN
 
 | | Human Aldose Reductase mutant T113S complexed with Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-01-15 | | Release date: | 2011-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
