4TWJ
 
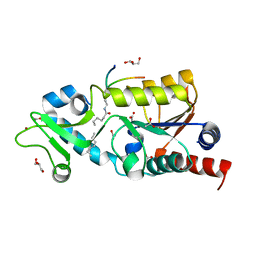 | | The structure of Sir2Af2 bound to a myristoylated histone peptide | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4 peptide, ... | | Authors: | Ringel, A.E, Roman, C, Wolberger, C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alternate deacylating specificities of the archaeal sirtuins Sir2Af1 and Sir2Af2.
Protein Sci., 23, 2014
|
|
4TWT
 
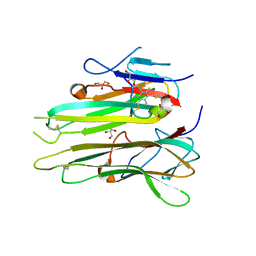 | | Human TNFa dimer in complex with the semi-synthetic bicyclic peptide M21 | | Descriptor: | (2,4,6-trimethylbenzene-1,3,5-triyl)trimethanol, ALA-CYS-PRO-PRO-CYS-LEU-TRP-GLN-VAL-LEU-CYS-GLY, GLYCEROL, ... | | Authors: | Luzi, S, Kondo, Y, Bernard, E, Stadler, L, Winter, G, Holliger, P. | | Deposit date: | 2014-07-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Subunit disassembly and inhibition of TNF alpha by a semi-synthetic bicyclic peptide.
Protein Eng.Des.Sel., 28, 2015
|
|
7ZOV
 
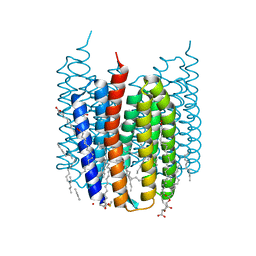 | | Crystal structure of Synechocystis halorhodopsin (SyHR), Cl-pumping mode, K state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, EICOSANE, ... | | Authors: | Kovalev, K, Bukhdruker, S, Astashkin, R, Vaganova, S, Gordeliy, V. | | Deposit date: | 2022-04-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into light-driven anion pumping in cyanobacteria.
Nat Commun, 13, 2022
|
|
5FTS
 
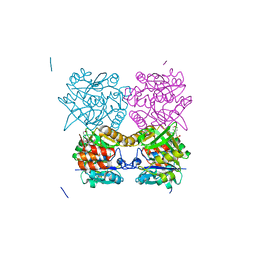 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Tran, F, Westwood, N.J, Naismith, J.H. | | Deposit date: | 2016-01-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric Competitive Inhibitors of the Glucose-1-Phosphate Thymidylyltransferase (Rmla) from Pseudomonas Aeruginosa.
To be Published
|
|
6GLC
 
 | | Structure of phospho-Parkin bound to phospho-ubiquitin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Gladkova, C, Maslen, S.L, Skehel, J.M, Komander, D. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of parkin activation by PINK1.
Nature, 559, 2018
|
|
4TV3
 
 | | Isolated p110a subunit of PI3Ka provides a platform for structure-based drug design | | Descriptor: | 2-amino-8-[trans-4-(2-hydroxyethoxy)cyclohexyl]-6-(6-methoxypyridin-3-yl)-4-methylpyrido[2,3-d]pyrimidin-7(8H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Deng, Y.-L, Bergqvist, S, Falk, M, Liu, W, Timofeevski, S. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Engineering of an isolated p110 alpha subunit of PI3K alpha permits crystallization and provides a platform for structure-based drug design.
Protein Sci., 23, 2014
|
|
6NPK
 
 | | Structure of the TM domain | | Descriptor: | Solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2019-08-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
6O8N
 
 | |
4TZT
 
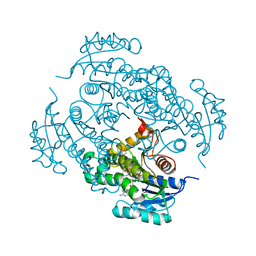 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ENOYL REDUCTASE (INHA) COMPLEXED WITH N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL- 5-OXOPYRROLIDINE-3-CARBOXAMIDE | | Descriptor: | (3S)-N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
5G09
 
 | | The crystal structure of a S-selective transaminase from Bacillus megaterium bound with R-alpha-methylbenzylamine | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, TETRAETHYLENE GLYCOL, ... | | Authors: | van Oosterwijk, N, Willies, S, Hekelaar, J, Terwisscha van Scheltinga, A.C, Turner, N.J, Dijkstra, B.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Substrate Range and Enantioselectivity of Two S-Selective Omega- Transaminases
Biochemistry, 55, 2016
|
|
4U1O
 
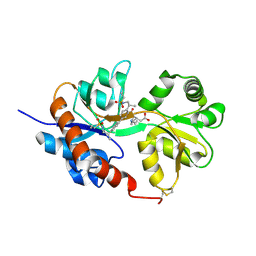 | | GluA2flip sLBD complexed with kainate and (R,R)-2b crystal form C | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, N,N'-[biphenyl-4,4'-diyldi(2R)propane-2,1-diyl]dipropane-2-sulfonamide | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
5DQC
 
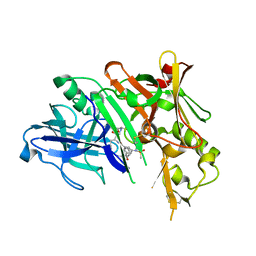 | | Co-crystal of BACE1 with compound 0211 | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-3-hydroxy-4-({(2S,3S)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Ghosh, A.K, Bhavanam, S.R, Yen, T.-C, Cardenas, E.L, Rao, K.V, Downs, D, Huang, X, Tang, J, Mescar, A.D. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-17 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.4651 Å) | | Cite: | Design of Potent and Highly Selective Inhibitors for Human beta-Secretase 2 (Memapsin 1), a Target for Type 2 Diabetes.
Chem Sci, 7, 2016
|
|
5DQZ
 
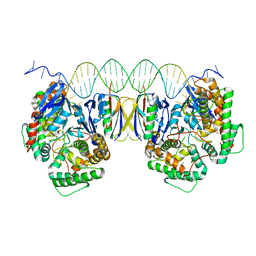 | | Crystal Structure of Cas-DNA-PAM complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (36-MER), ... | | Authors: | Wang, J, Li, J, Zhao, H, Sheng, G, Wang, M, Yin, M, Wang, Y. | | Deposit date: | 2015-09-15 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems.
Cell, 163, 2015
|
|
6FEW
 
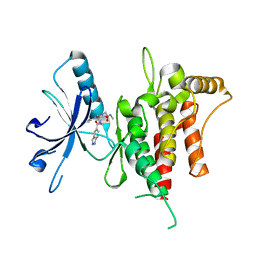 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.440A, P1211, Rfree=24.1% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6OVX
 
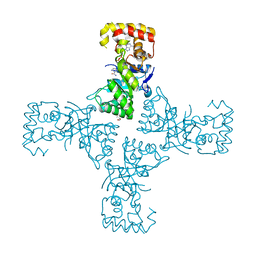 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+, P422 form | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4U38
 
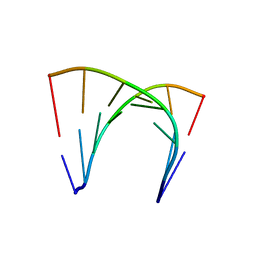 | | RNA duplex containing UU mispair | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
6NF2
 
 | |
4UBS
 
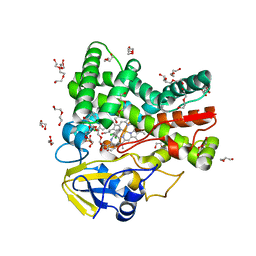 | | The crystal structure of cytochrome P450 105D7 from Streptomyces avermitilis in complex with Diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Xu, L.H, Ikeda, H, Arakawa, T, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2014-08-13 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the 4'-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
5EBM
 
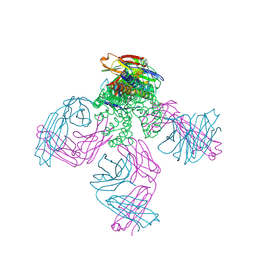 | | KcsA T75G mutant in the nonconductive state | | Descriptor: | Antibody Fab Fragment Light Chain, DIACYL GLYCEROL, NONAN-1-OL, ... | | Authors: | Matulef, K, Valiyaveetil, F.I. | | Deposit date: | 2015-10-19 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-type Inactivation and in Recovery from Inactivation.
Structure, 24, 2016
|
|
6NS8
 
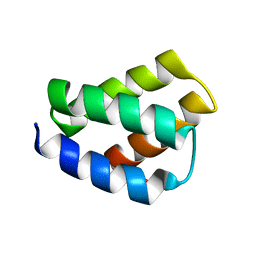 | | RDC-refined SOLUTION NMR STRUCTURE OF PROTEIN PF2048.1 | | Descriptor: | Uncharacterized protein | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
6NT3
 
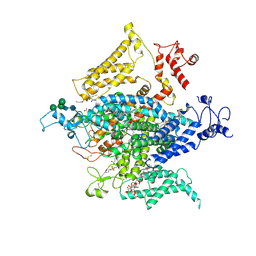 | | Cryo-EM structure of a human-cockroach hybrid Nav channel. | | Descriptor: | (7E,21R,24S)-27-amino-24-hydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaheptacos-7-en-21-yl (9Z,12E)-octadeca-9,12-dienoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clairfeuille, T, Rohou, A, Payandeh, J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-02-20 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a human-cockroach hybrid Nav channel in the presence and absence of the alpha-scorpion toxin AaH2.
Science, 2019
|
|
5E25
 
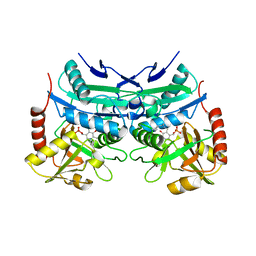 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Geoglobus acetivorans complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, PYRIDOXAL-5'-PHOSPHATE, branched-chain aminotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
4U61
 
 | |
4U60
 
 | |
4U77
 
 | | BTB domain from Drosophila CP190 | | Descriptor: | Centrosome-associated zinc finger protein CP190, DIMETHYL SULFOXIDE, GLYCEROL | | Authors: | Allemand, F, Cohen-Gonsaud, M, Labesse, G, Nollmann, M. | | Deposit date: | 2014-07-30 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome.
Plos Genet., 10, 2014
|
|
