8A4M
 
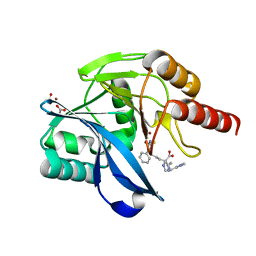 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in complex with compound 8 (JMV-7061) | | Descriptor: | (2~{S})-2-[bis(1~{H}-imidazol-4-ylmethyl)amino]-5-(3-phenyl-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl)pentanoic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Corsica, G, Sannio, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of 1,2,4-Triazole-3-thiones toward Broad-Spectrum Metallo-beta-lactamase Inhibitors Showing Potent Synergistic Activity on VIM- and NDM-1-Producing Clinical Isolates.
J.Med.Chem., 65, 2022
|
|
4RYW
 
 | |
8U1S
 
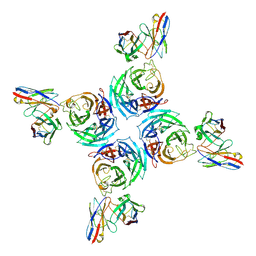 | |
6N0D
 
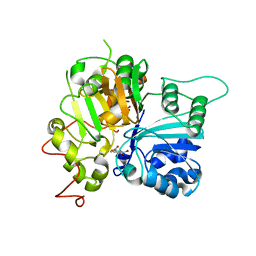 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ575 | | Descriptor: | 1,2-ETHANEDIOL, 4-fluorobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain in complex with compound XZ575
To Be Published
|
|
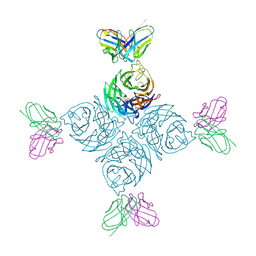
8U1Q
 
 | |
8XW6
 
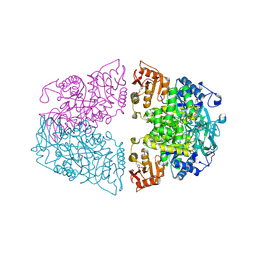 | | Crystal structure of Streptococcus pneumoniae pyruvate kinase in complex with oxalate and fructose 1,6-bisphosphate and ATP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nakashima, R, Taguchi, A. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Nucleotide Selectivity in Pyruvate Kinase.
J.Mol.Biol., 436, 2024
|
|
6MUL
 
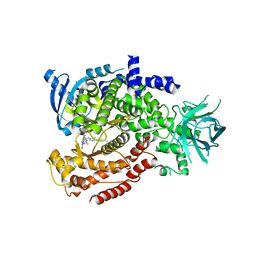 | | Murine PI3K delta kinsae domain - cpd 1 | | Descriptor: | 1-{1-[8-(1-ethyl-5-methyl-1H-pyrazol-4-yl)-9-methyl-9H-purin-6-yl]piperidin-4-yl}-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Fischmann, T.O. | | Deposit date: | 2018-10-23 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure Overhaul Affords a Potent Purine PI3K delta Inhibitor with Improved Tolerability.
J.Med.Chem., 62, 2019
|
|
6GH0
 
 | |
5DFG
 
 | |
7ZUA
 
 | |
6GON
 
 | | Crystal Structure Of Sea Bream Transthyretin in complex with Perfluorooctanoic acid (PFOA). Crystallized in PEG | | Descriptor: | Transthyretin, pentadecafluorooctanoic acid | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
4S2V
 
 | | E. coli RppH structure, KI soak | | Descriptor: | ACETATE ION, CALCIUM ION, IODIDE ION, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
5DII
 
 | | Structure of an engineered bacterial microcompartment shell protein binding a [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, Microcompartments protein | | Authors: | Sutter, M, Aussignargues, C, Turmo, A, Kerfeld, C.A. | | Deposit date: | 2015-09-01 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster.
J.Am.Chem.Soc., 138, 2016
|
|
6N8F
 
 | |
6N2J
 
 | | Tetrahydropyridopyrimidines as Covalent Inhibitors of KRAS-G12C | | Descriptor: | 1-{4-[7-(naphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P. | | Deposit date: | 2018-11-13 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity.
ACS Med Chem Lett, 9, 2018
|
|
6NV2
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 in complex with DP005 | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-02-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
6NB3
 
 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6QJ0
 
 | |
4SGA
 
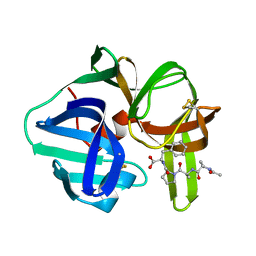 | |
6QK2
 
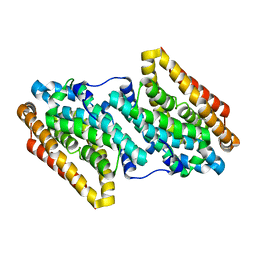 | |
6OR0
 
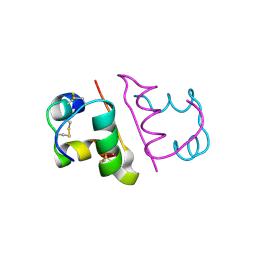 | | Crystal structure of Insulin from Non-merohedrally twinned crystals | | Descriptor: | Insulin chain A, Insulin chain B | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4TXV
 
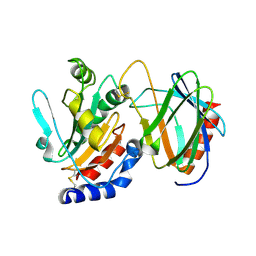 | | Crystal structure of the mixed disulfide intermediate between thioredoxin-like TlpAs(C110S) and subunit II of cytochrome c oxidase CoxBPD (C233S) | | Descriptor: | Cytochrome c oxidase subunit 2, Thiol:disulfide interchange protein TlpA | | Authors: | Quade, N, Abicht, H.K, Hennecke, H, Glockshuber, R. | | Deposit date: | 2014-07-07 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How Periplasmic Thioredoxin TlpA Reduces Bacterial Copper Chaperone ScoI and Cytochrome Oxidase Subunit II (CoxB) Prior to Metallation.
J.Biol.Chem., 289, 2014
|
|
6NCS
 
 | |
7ZPH
 
 | | Crystal structure of Pizza6-TNK-RSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TNK-RSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
6GX3
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with an hydroxamate 1 | | Descriptor: | 4-chloranyl-~{N}-oxidanyl-1-benzothiophene-2-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, Crystallization Studies, and in vitro Characterization of Cinnamic Acid Derivatives as SmHDAC8 Inhibitors for the Treatment of Schistosomiasis.
ChemMedChem, 13, 2018
|
|
