1VXE
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXD
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1DOI
 
 | | 2FE-2S FERREDOXIN FROM HALOARCULA MARISMORTUI | | Descriptor: | 2FE-2S FERREDOXIN, FE2/S2 (INORGANIC) CLUSTER, POTASSIUM ION | | Authors: | Frolow, F, Harel, M, Sussman, J.L, Shoham, M. | | Deposit date: | 1996-04-08 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into protein adaptation to a saturated salt environment from the crystal structure of a halophilic 2Fe-2S ferredoxin.
Nat.Struct.Biol., 3, 1996
|
|
1VXG
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXF
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXH
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1VXC
 
 | | NATIVE SPERM WHALE MYOGLOBIN | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, F, Phillips Jr, G.N. | | Deposit date: | 1996-01-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of CO-, deoxy- and met-myoglobins at various pH values.
J.Mol.Biol., 256, 1996
|
|
1NOI
 
 | |
1CQG
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN REF-1 (RESIDUES 59-71 OF THE P50 SUBUNIT OF NFKB), NMR, 31 STRUCTURES | | Descriptor: | REF-1 PEPTIDE, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1996-04-02 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal.
Structure, 4, 1996
|
|
1CQH
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN REF-1 (RESIDUES 59-71 OF THE P50 SUBUNIT OF NFKB), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REF-1 PEPTIDE, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1996-04-02 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal.
Structure, 4, 1996
|
|
1CLZ
 
 | |
1MZM
 
 | |
1DNP
 
 | | STRUCTURE OF DEOXYRIBODIPYRIMIDINE PHOTOLYASE | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Park, H.-W, Sancar, A, Deisenhofer, J. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of DNA photolyase from Escherichia coli.
Science, 268, 1995
|
|
1PYC
 
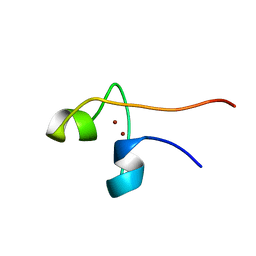 | | CYP1 (HAP1) DNA-BINDING DOMAIN (RESIDUES 60-100), NMR, 15 STRUCTURES | | Descriptor: | CYP1, ZINC ION | | Authors: | Timmerman, J, Vuidepot, A.-L, Bontems, F, Lallemand, J.-Y, Gervais, M, Shechter, E, Guiard, B. | | Deposit date: | 1996-02-17 | | Release date: | 1996-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N resonance assignment and three-dimensional structure of CYP1 (HAP1) DNA-binding domain.
J.Mol.Biol., 259, 1996
|
|
1UMU
 
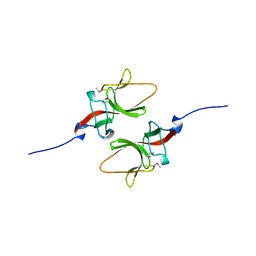 | |
2ASI
 
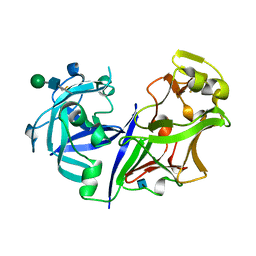 | | ASPARTIC PROTEINASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARTIC PROTEINASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Jia, Z, Vandonselaar, M, Kepliakov, P.S.A, Quail, J.W. | | Deposit date: | 1995-12-09 | | Release date: | 1996-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the aspartic proteinase from Rhizomucor miehei at 2.15 A resolution.
J.Mol.Biol., 268, 1997
|
|
1CMR
 
 | | NMR SOLUTION STRUCTURE OF A CHIMERIC PROTEIN, DESIGNED BY TRANSFERRING A FUNCTIONAL SNAKE BETA-HAIRPIN INTO A SCORPION ALPHA/BETA SCAFFOLD (PH 3.5, 20C), NMR, 18 STRUCTURES | | Descriptor: | CHARYBDOTOXIN, ALPHA CHIMERA | | Authors: | Zinn-Justin, S, Guenneugues, M, Drakopoulou, E, Gilquin, B, Vita, C, Menez, A. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Transfer of a beta-hairpin from the functional site of snake curaremimetic toxins to the alpha/beta scaffold of scorpion toxins: three-dimensional solution structure of the chimeric protein.
Biochemistry, 35, 1996
|
|
1FUG
 
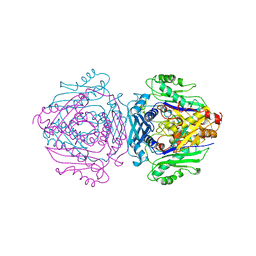 | | S-ADENOSYLMETHIONINE SYNTHETASE | | Descriptor: | S-ADENOSYLMETHIONINE SYNTHETASE | | Authors: | Fu, Z, Markham, G.D, Takusagawa, F. | | Deposit date: | 1996-02-25 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Flexible loop in the structure of S-adenosylmethionine synthetase crystallized in the tetragonal modification.
J.Biomol.Struct.Dyn., 13, 1996
|
|
1ARL
 
 | | CARBOXYPEPTIDASE A WITH ZN REMOVED | | Descriptor: | APO-CARBOXYPEPTIDASE A=ALPHA= (COX) | | Authors: | Greenblatt, H.M, Feinberg, H, Tucker, P.A, Shoham, G. | | Deposit date: | 1994-11-22 | | Release date: | 1996-08-01 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1XTC
 
 | | CHOLERA TOXIN | | Descriptor: | CHOLERA TOXIN | | Authors: | Zhang, R.-G, Westbrook, E. | | Deposit date: | 1996-01-10 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional crystal structure of cholera toxin.
J.Mol.Biol., 251, 1995
|
|
2ORA
 
 | | RHODANESE (THIOSULFATE: CYANIDE SULFURTRANSFERASE) | | Descriptor: | OXIDIZED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
1TUC
 
 | |
1DMO
 
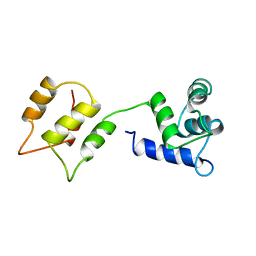 | | CALMODULIN, NMR, 30 STRUCTURES | | Descriptor: | CALMODULIN | | Authors: | Zhang, M, Tanaka, T, Ikura, M. | | Deposit date: | 1996-04-24 | | Release date: | 1996-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced conformational transition revealed by the solution structure of apo calmodulin.
Nat.Struct.Biol., 2, 1995
|
|
1IGB
 
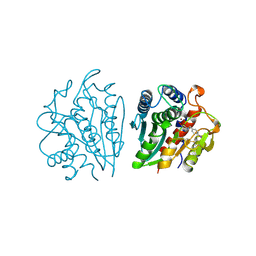 | | AEROMONAS PROTEOLYTICA AMINOPEPTIDASE COMPLEXED WITH THE INHIBITOR PARA-IODO-D-PHENYLALANINE HYDROXAMATE | | Descriptor: | AMINOPEPTIDASE, PARA-IODO-D-PHENYLALANINE HYDROXAMIC ACID, ZINC ION | | Authors: | Chevrier, B, D'Orchymont, H, Schalk, C, Tarnus, C, Moras, D. | | Deposit date: | 1996-02-27 | | Release date: | 1996-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the Aeromonas proteolytica aminopeptidase complexed with a hydroxamate inhibitor. Involvement in catalysis of Glu151 and two zinc ions of the co-catalytic unit.
Eur.J.Biochem., 237, 1996
|
|
1CLY
 
 | |
