7V15
 
 | | Factor XIa in Complex with Compound 2i | | Descriptor: | 5-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]-4-methyl-1,3-thiazole, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Cedervall, P, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
6A0C
 
 | | Structure of a triple-helix region of human collagen type III | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, collagen type III peptide | | Authors: | Yang, X, Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Characterization by high-resolution crystal structure analysis of a triple-helix region of human collagen type III with potent cell adhesion activity.
Biochem. Biophys. Res. Commun., 508, 2019
|
|
7V10
 
 | | Factor XIa in Complex with Compound 2d | | Descriptor: | CITRIC ACID, Coagulation factor XIa light chain, methyl ~{N}-[4-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]phenyl]carbamate | | Authors: | Shaffer, P.L, Spurlino, J, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
7UGQ
 
 | |
8PCH
 
 | | CRYSTAL STRUCTURE OF PORCINE CATHEPSIN H DETERMINED AT 2.1 ANGSTROM RESOLUTION: LOCATION OF THE MINI-CHAIN C-TERMINAL CARBOXYL GROUP DEFINES CATHEPSIN H AMINOPEPTIDASE FUNCTION | | Descriptor: | CATHEPSIN H, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guncar, G, Podobnik, M, Pungercar, J, Strukelj, B, Turk, V, Turk, D. | | Deposit date: | 1997-11-07 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of porcine cathepsin H determined at 2.1 A resolution: location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function.
Structure, 6, 1998
|
|
7UPY
 
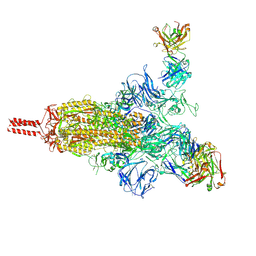 | | An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
8P9G
 
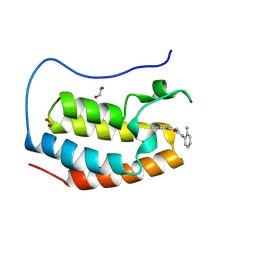 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB390 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[(~{E})-(6-methyl-7-oxidanyl-1~{H}-indol-4-yl)diazenyl]benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9H
 
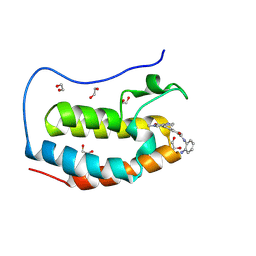 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB437 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-2-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)quinoline-6-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9I
 
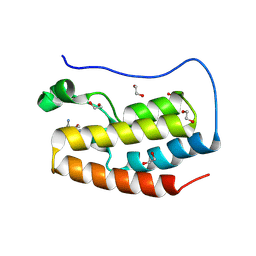 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB462 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9K
 
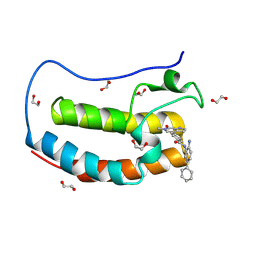 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB503 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
7UOH
 
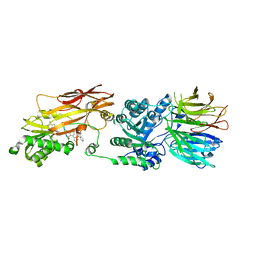 | | PRMT5/MEP50 crystal structure with MTA and an achiral, class 1, non-atropisomeric inhibitor bound | | Descriptor: | (2M)-2-[(4M)-4-{4-(aminomethyl)-1-oxo-8-[(2R)-oxolan-2-yl]-1,2-dihydrophthalazin-6-yl}-1-methyl-1H-pyrazol-5-yl]-1-benzothiophene-3-carbonitrile, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and evaluation of achiral, non-atropisomeric 4-(aminomethyl)phthalazin-1(2H)-one derivatives as novel PRMT5/MTA inhibitors.
Bioorg.Med.Chem., 71, 2022
|
|
8P9J
 
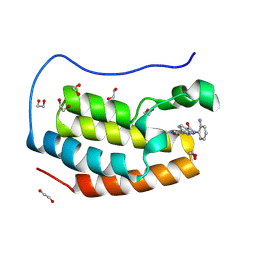 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB500 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8PDF
 
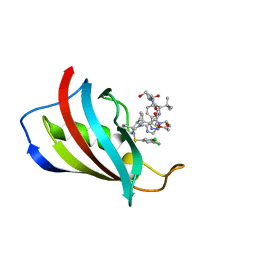 | | FKBP12 in complex with PROTAC 6a2 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[4-[(1~{S})-1-[(1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-2-oxidanylidene-3,10-diazabicyclo[4.3.1]decan-3-yl]ethyl]-1,2,3-triazol-1-yl]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Walz, M, Geiger, T.M, Hausch, F. | | Deposit date: | 2023-06-12 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a Potent Proteolysis Targeting Chimera Enables Targeting the Scaffolding Functions of FK506-Binding Protein 51 (FKBP51).
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7UCG
 
 | |
8DVD
 
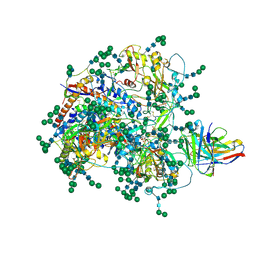 | |
6AFW
 
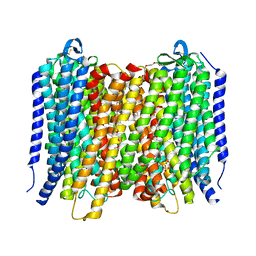 | | Proton pyrophosphatase-T228D mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
7UU5
 
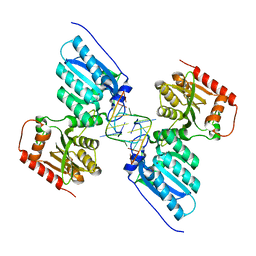 | | Crystal structure of APOBEC3G complex with 5'-Overhang dsRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*CP*CP*GP*CP*AP*GP*CP*G)-3'), RNA (5'-R(P*UP*AP*AP*CP*GP*CP*UP*GP*CP*GP*G)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
8DO5
 
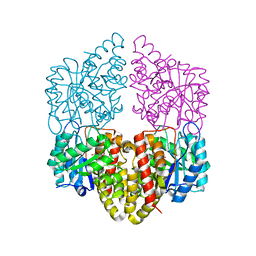 | | Crystal structure of NahE in complex with intermediate (R)-4-hydroxy-4-(2-hydroxyphenyl)-2-iminobutanoate | | Descriptor: | (4R)-4-hydroxy-4-(2-hydroxyphenyl)butanoic acid, DIMETHYL SULFOXIDE, Trans-ohydrobenzylidenepyruvate hydratase aldolase | | Authors: | LeVieux, J.A, Hardtke, H.A, Zhang, Y.J. | | Deposit date: | 2022-07-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mutagenic analysis of NahE, a hydratase-aldolase in the naphthalene degradative pathway.
Arch.Biochem.Biophys., 733, 2023
|
|
6AFT
 
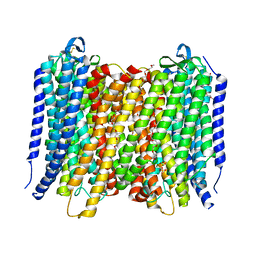 | | Proton pyrophosphatase - E301Q | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Tang, K.-Z, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AC0
 
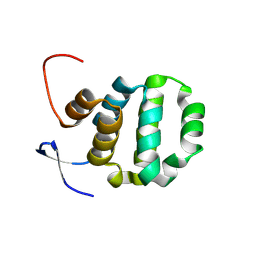 | | Crystal structure of TRADD death domain GlcNAcylated by EPEC effector NleB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor type 1-associated DEATH domain protein | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural and Functional Insights into Host Death Domains Inactivation by the Bacterial Arginine GlcNAcyltransferase Effector.
Mol.Cell, 74, 2019
|
|
6AFV
 
 | | Proton pyrophosphatase-L555K mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6ADB
 
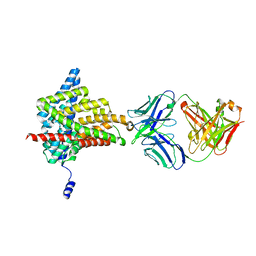 | | Crystal structure of the E148N mutant CLC-ec1 in 20mM bromide | | Descriptor: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment, ... | | Authors: | Lim, H.-H, Park, K. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6AL1
 
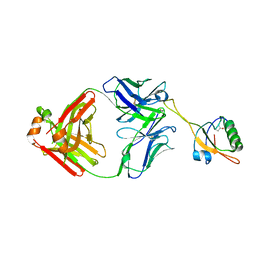 | | The NZ-1 Fab complexed with the PDZ tandem fragment of A. aeolicus S2P homolog with the PA12 tag inserted between the residues 181 and 184 | | Descriptor: | Heavy chain of antigen binding fragment, Fab of NZ-1, Light chain of antigen binding fragment, ... | | Authors: | Tamura, R, Oi, R, Kaneko, M.K, Kato, Y, Nogi, T. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins.
Protein Sci., 28, 2019
|
|
6AFU
 
 | | Proton pyrophosphatase-L555M mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
