1D2L
 
 | |
1D2M
 
 | | UVRB PROTEIN OF THERMUS THERMOPHILUS HB8; A NUCLEOTIDE EXCISION REPAIR ENZYME | | Descriptor: | EXCINUCLEASE ABC SUBUNIT B, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Nakagawa, N, Sugahara, M, Masui, R, Kato, R, Fukuyama, K, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-09-25 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair.
J.Biochem.(Tokyo), 126, 1999
|
|
1D2N
 
 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
1D2O
 
 | |
1D2P
 
 | |
1D2Q
 
 | | CRYSTAL STRUCTURE OF HUMAN TRAIL | | Descriptor: | TNF-RELATED APOPTOSIS INDUCING LIGAND | | Authors: | Cha, S.-S. | | Deposit date: | 1999-09-27 | | Release date: | 2000-02-11 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity.
Immunity, 11, 1999
|
|
1D2R
 
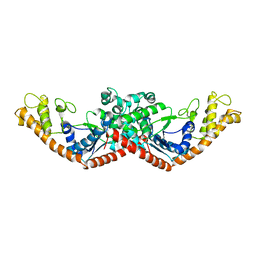 | |
1D2S
 
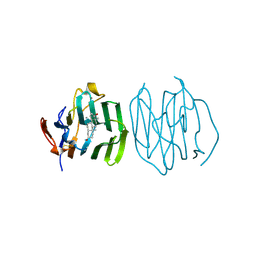 | | CRYSTAL STRUCTURE OF THE N-TERMINAL LAMININ G-LIKE DOMAIN OF SHBG IN COMPLEX WITH DIHYDROTESTOSTERONE | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, CALCIUM ION, SEX HORMONE-BINDING GLOBULIN | | Authors: | Grishkovskaya, I, Avvakumov, G.V, Sklenar, G, Dales, D, Hammond, G.L, Muller, Y.A. | | Deposit date: | 1999-09-28 | | Release date: | 2000-06-28 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human sex hormone-binding globulin: steroid transport by a laminin G-like domain.
EMBO J., 19, 2000
|
|
1D2T
 
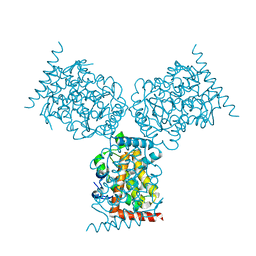 | | CRYSTAL STRUCTURE OF ACID PHOSPHATASE FROM ESCHERICHIA BLATTAE | | Descriptor: | ACID PHOSPHATASE, SULFATE ION | | Authors: | Ishikawa, K, Mihara, Y, Gondoh, K, Suzuki, E, Asano, Y. | | Deposit date: | 1999-09-28 | | Release date: | 2000-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate.
EMBO J., 19, 2000
|
|
1D2U
 
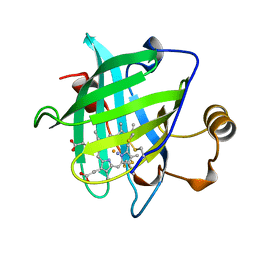 | | 1.15 A CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Andersen, J.F, Roberts, S.A, Montfort, W.R. | | Deposit date: | 1999-09-28 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1D2V
 
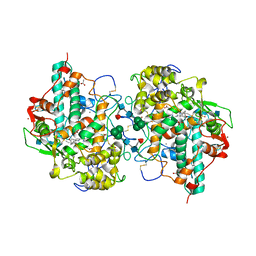 | | CRYSTAL STRUCTURE OF BROMIDE-BOUND HUMAN MYELOPEROXIDASE ISOFORM C AT PH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BROMIDE ION, ... | | Authors: | Fiedler, T.J, Davey, C.A, Fenna, R.E. | | Deposit date: | 1999-09-28 | | Release date: | 2000-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure and characterization of halide-binding sites of human myeloperoxidase at 1.8 A resolution.
J.Biol.Chem., 275, 2000
|
|
1D2W
 
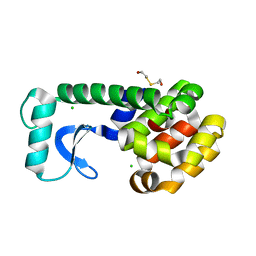 | | N-TERMINAL DOMAIN CORE METHIONINE MUTATION | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Matthews, B.W. | | Deposit date: | 1999-09-28 | | Release date: | 1999-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Use of differentially substituted selenomethionine proteins in X-ray structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1D2Y
 
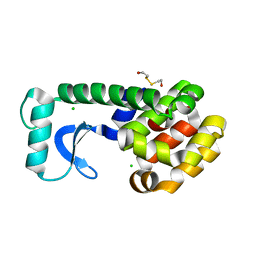 | | N-TERMINAL DOMAIN CORE METHIONINE MUTATION | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Matthews, B.W. | | Deposit date: | 1999-09-28 | | Release date: | 1999-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Use of differentially substituted selenomethionine proteins in X-ray structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1D2Z
 
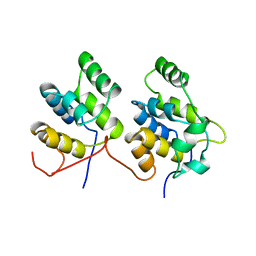 | | THREE-DIMENSIONAL STRUCTURE OF A COMPLEX BETWEEN THE DEATH DOMAINS OF PELLE AND TUBE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DEATH DOMAIN OF PELLE, DEATH DOMAIN OF TUBE | | Authors: | Xiao, T, Towb, P, Wasserman, S.A, Sprang, S.R. | | Deposit date: | 1999-09-28 | | Release date: | 1999-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a complex between the death domains of Pelle and Tube.
Cell(Cambridge,Mass.), 99, 1999
|
|
1D30
 
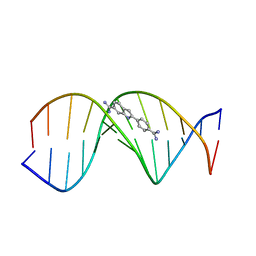 | | THE STRUCTURE OF DAPI BOUND TO DNA | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Larsen, T, Goodsell, D.S, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1991-01-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of DAPI bound to DNA.
J.Biomol.Struct.Dyn., 7, 1989
|
|
1D31
 
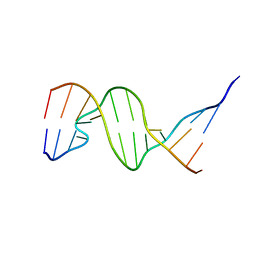 | | THE THREE-DIMENSIONAL STRUCTURES OF BULGE-CONTAINING DNA FRAGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Joshua-Tor, L, Frolow, F, Appella, E, Hope, H, Rabinovich, D, Sussman, J.L. | | Deposit date: | 1991-04-25 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures of bulge-containing DNA fragments.
J.Mol.Biol., 225, 1992
|
|
1D32
 
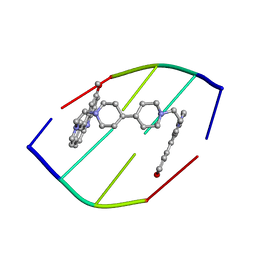 | | DRUG-INDUCED DNA REPAIR: X-RAY STRUCTURE OF A DNA-DITERCALINIUM COMPLEX | | Descriptor: | DITERCALINIUM, DNA (5'-D(*CP*GP*CP*G)-3') | | Authors: | Gao, Q, Williams, L.D, Egli, M, Rabinovich, D, Chen, S.-L, Quigley, G.J, Rich, A. | | Deposit date: | 1991-01-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Drug-induced DNA repair: X-ray structure of a DNA-ditercalinium complex.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D33
 
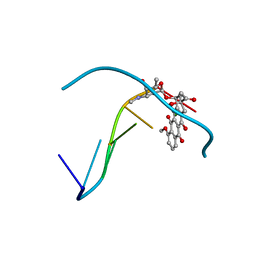 | | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-RAY diffraction studies | | Descriptor: | 5'-D(*CP*GP*CP*(G49)P*CP*G)-3', DAUNOMYCIN, MAGNESIUM ION | | Authors: | Wang, A.H.-J, Gao, Y.-G, Liaw, Y.-C, Li, Y.-K. | | Deposit date: | 1991-02-27 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Formaldehyde cross-links daunorubicin and DNA efficiently: HPLC and X-ray diffraction studies.
Biochemistry, 30, 1991
|
|
1D35
 
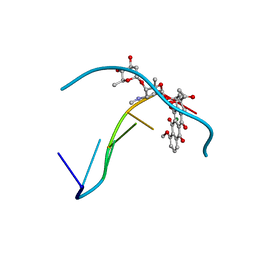 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(A40)P*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D36
 
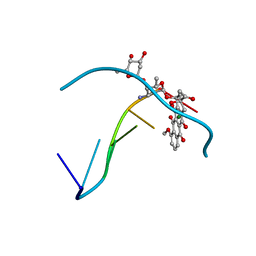 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1D37
 
 | |
1D38
 
 | |
1D39
 
 | | COVALENT MODIFICATION OF GUANINE BASES IN DOUBLE STRANDED DNA: THE 1.2 ANGSTROMS Z-DNA STRUCTURE OF D(CGCGCG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) ION, DNA (5'-D(*CP*(CU)GP*CP*(CU)GP*CP*(CU)G)-3'), SODIUM ION | | Authors: | Kagawa, T.F, Geierstanger, B.H, Wang, A.H.-J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Covalent modification of guanine bases in double-stranded DNA. The 1.2-A Z-DNA structure of d(CGCGCG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
1D3A
 
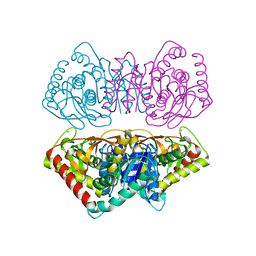 | | CRYSTAL STRUCTURE OF THE WILD TYPE HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, HALOPHILIC MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-09-28 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
1D3B
 
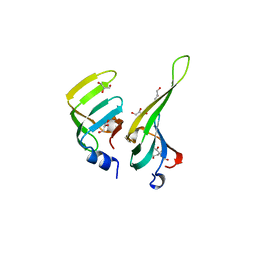 | | CRYSTAL STRUCTURE OF THE D3B SUBCOMPLEX OF THE HUMAN CORE SNRNP DOMAIN AT 2.0A RESOLUTION | | Descriptor: | CITRIC ACID, GLYCEROL, PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN ASSOCIATED PROTEIN B), ... | | Authors: | Kambach, C, Walke, S, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
