7OVF
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 8 (JMV-7207) | | Descriptor: | 4-[2-[(4-fluorophenyl)methylsulfanyl]ethyl]-3-phenyl-1H-1,2,4-triazole-5-thione, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Marcoccia, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7OJQ
 
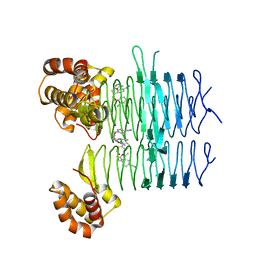 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 7 | | Descriptor: | 2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]-~{N}-[(5-methyl-1,3,4-oxadiazol-2-yl)methyl]ethanamide, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OJW
 
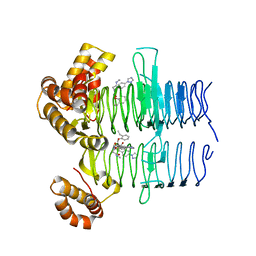 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 93 | | Descriptor: | 2-[2-(2-ethylphenoxy)ethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-~{N}-methyl-ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7P3V
 
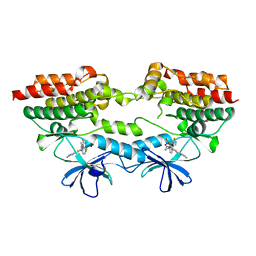 | | B-Raf V600E structure bound to a new inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, ~{N}-[3-[5-(2-azanylpyrimidin-4-yl)-2-[(3~{S})-morpholin-3-yl]-1,3-thiazol-4-yl]-2-fluoranyl-phenyl]-2,5-bis(fluoranyl)benzenesulfonamide | | Authors: | Schneider, M, Gelin, M, Cohen-Gonsaud, M, Labesse, G. | | Deposit date: | 2021-07-08 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based and Knowledge-Informed Design of B-Raf Inhibitors Devoid of Deleterious PXR Binding.
J.Med.Chem., 65, 2022
|
|
7OKD
 
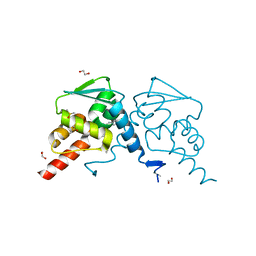 | | Crystal structure of human BCL6 BTB domain in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[[4-[2-(5-cyclopropylpyrimidin-2-yl)propan-2-ylamino]-1-methyl-2-oxidanylidene-quinolin-6-yl]amino]pyridine-3-carbonitrile, B-cell lymphoma 6 protein, ... | | Authors: | Gunnell, E.A, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
J.Med.Chem., 64, 2021
|
|
7OLY
 
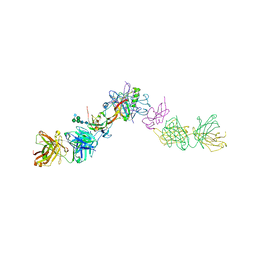 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OTB
 
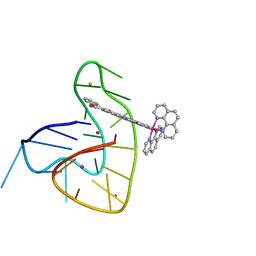 | | Ruthenium polypridyl complex bound to a unimolecular chair-form G-quadruplex | | Descriptor: | BARIUM ION, DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), POTASSIUM ION, ... | | Authors: | McQuaid, K.T, Cardin, C.J, Hall, J.P, Paterson, N.G, Baumgaertner, L. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ruthenium Polypyridyl Complex Bound to a Unimolecular Chair-Form G-Quadruplex.
J.Am.Chem.Soc., 144, 2022
|
|
7OHO
 
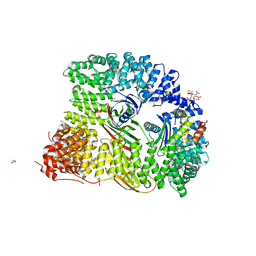 | | Crystal structure of AP2 FCHO2 chimera | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta,F-BAR domain only protein 2, AP-2 complex subunit mu, ... | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
7OLI
 
 | | Crystal structure of Pab-AGOG in complex with 8-oxoguanosine | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXY-8-OXOGUANOSINE, N-glycosylase/DNA lyase | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OKZ
 
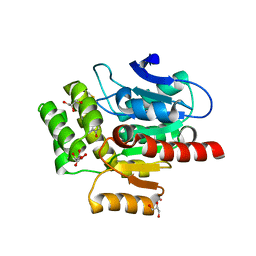 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH 2-METHYL- QUINOLIN-4(1H)-ONE UNDER HYPEROXIC CONDITIONS | | Descriptor: | 1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase, 2-methyl-quinolin-4(1H)-one, D(-)-TARTARIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2021-05-18 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha / beta-hydrolase fold.
Chem Sci, 14, 2023
|
|
7OOM
 
 | |
7OOU
 
 | | NaK C-DI mutant with Li+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OVS
 
 | | Heterodimeric murine tRNA-guanine transglycosylase in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
7OY0
 
 | | Structure of human Spermine Oxidase in complex with a highly selective allosteric inhibitor | | Descriptor: | 4-[(4-imidazo[1,2-a]pyridin-3-yl-1,3-thiazol-2-yl)amino]phenol, CHLORIDE ION, FAD-MDL72527 adduct, ... | | Authors: | Impagliazzo, A, Thomsen, M, Johannsson, S, Krapp, S. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
7OY7
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing cytosine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OWZ
 
 | | Heterodimeric murine tRNA-guanine transglycosylase in complex with queuine and in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
7P8L
 
 | | Crystal structure of Pyrococcus abyssi 8-oxoguanine DNA glycosylase (PabAGOG) in complex with dsDNA containing cytosine opposite to 8-oxoG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(8OG)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Flament, D, Castaing, B. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P9Z
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing adenine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P9Y
 
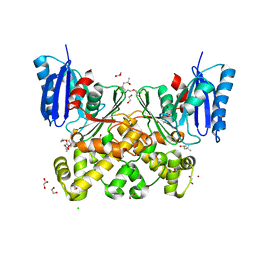 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7OPV
 
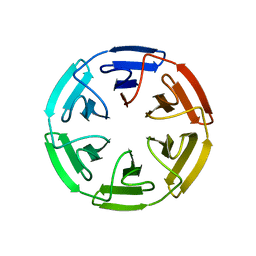 | | Crystal structure of the computationally designed SAKe6BE-3HH protein, alternative packing | | Descriptor: | SAKe6BE-3HH | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-06-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7OP4
 
 | |
7OXF
 
 | |
