5J5Z
 
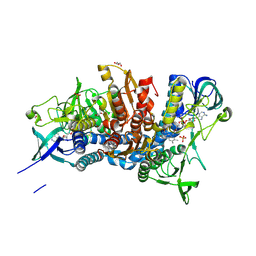 | | Crystal structure of the D444V disease-causing mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Mizsei, R, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2016-04-04 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase.
Free Radic. Biol. Med., 124, 2018
|
|
5LPP
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex (space group C2) with 6-amino-4-(2-((3aR,4R,6R,6aR)-6-methoxy-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)ethyl)-2-(methylamino)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 4-[2-[(3~{a}~{R},4~{R},6~{R},6~{a}~{R})-4-methoxy-2,2-dimethyl-3~{a},4,6,6~{a}-tetrahydrofuro[3,4-d][1,3]dioxol-6-yl]ethyl]-6-azanyl-2-(methylamino)-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
8JM5
 
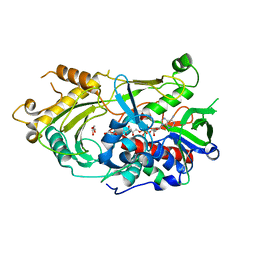 | | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A/S333V/P340L from Prunus communis | | Descriptor: | (R)-mandelonitrile lyase, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Y.-C, Li, F.-L, Yu, H.-L, Xu, J.-H. | | Deposit date: | 2023-06-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Endo-deglycosylated hydroxynitrile lyase isozyme 5 mutant L331A/S333V/P340L from Prunus communis
To Be Published
|
|
9C8C
 
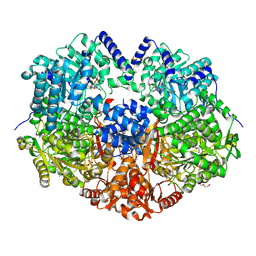 | |
9C0T
 
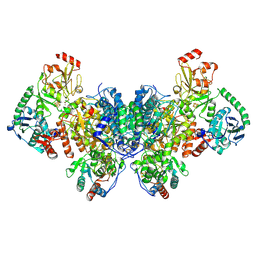 | |
5N56
 
 | |
8YFM
 
 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS122 | | Descriptor: | GLYCEROL, RB1-inducible coiled-coil protein 1, SULFATE ION, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFN
 
 | |
4UCF
 
 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, N-PROPANOL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpha-Galactose.
FEBS J., 283, 2016
|
|
5HXQ
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with DMSPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of a Z,Z-Farnesyl Diphosphate Synthase
Acs Omega, 2, 2017
|
|
8T1W
 
 | | Crystal structure of orphan G protein-coupled receptor 6 with bound CVN424 | | Descriptor: | 1-{2-[4-(2,4-difluorophenoxy)piperidin-1-yl]-3-{[(3R)-oxolan-3-yl]amino}-7,8-dihydropyrido[3,4-b]pyrazin-6(5H)-yl}ethan-1-one, G-protein coupled receptor 6, Soluble cytochrome b562 chimera | | Authors: | Barekatain, M, Johansson, L, Lam, J.H, Sadybekov, A.V, Han, G.W, Popov, P, Russo, J, Bliesath, J, Brice, N, Beresford, M, Carlson, L, Saikatendu, K.S, Sun, H, Murphy, S, Monenschein, H, Schiffer, H.H, Lutomski, C, Robinson, C.V, Liu, Z, Hua, T, Katritch, V, Cherezov, V. | | Deposit date: | 2023-06-04 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structural insights into the high basal activity and inverse agonism of the orphan receptor GPR6 implicated in Parkinson's disease.
Sci.Signal., 17, 2024
|
|
5LM3
 
 | | Plasmodium falciparum nicotinic acid mononucleotide adenylyltransferase complexed with APC | | Descriptor: | 1,2-ETHANEDIOL, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Nicotinate-nucleotide adenylyltransferase | | Authors: | Bathke, J, Fritz-Wolf, K, Rahlfs, S, Brandstaether, C, Burkhardt, A, Becker, K. | | Deposit date: | 2016-07-29 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Plasmodium falciparum Nicotinic Acid Mononucleotide Adenylyltransferase.
J. Mol. Biol., 428, 2016
|
|
5HPI
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain-3-Hydroxy Benzoic Acid complex from Acinetobacter | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, SULFATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-09-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
6RPQ
 
 | | Crystal structure of PhoCDC21-1 intein | | Descriptor: | Ubiquitin-like protein SMT3,1108aa long hypothetical cell division control protein | | Authors: | Beyer, H.M, Mikula, K.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
5LGZ
 
 | | Structure of Photoreduced Pentaerythritol Tetranitrate Reductase | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ISOPROPYL ALCOHOL, Pentaerythritol tetranitrate reductase | | Authors: | Kwon, H, Smith, O.M, Moody, P.C.E. | | Deposit date: | 2016-07-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combining X-ray and neutron crystallography with spectroscopy.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5HXT
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with IPP and DMSPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of a Z,Z-Farnesyl Diphosphate Synthase
Acs Omega, 2, 2017
|
|
6EPW
 
 | | The ATAD2 bromodomain in complex with compound UZH-DU32 | | Descriptor: | (2~{R})-~{N}-[5-[3,5-bis(oxidanyl)phenyl]-4-ethanoyl-1,3-thiazol-2-yl]-2-carbamimidamido-propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | Hitting a moving target: simulation and crystallography study of ATAD2 bromodomain blockers
To Be Published
|
|
6EPJ
 
 | | The ATAD2 bromodomain in complex with compound 6 | | Descriptor: | (2~{R})-2-azanyl-~{N}-[4-ethanoyl-5-(3-hydroxyphenyl)-1,3-thiazol-2-yl]propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
9N4U
 
 | | Crystal structure of PAK1 bound to compound R1 | | Descriptor: | (3M)-3-[(4P)-2-chloro-4-(6-methylpyridin-2-yl)phenyl]-1-{2-[2-(dimethylamino)ethoxy]ethyl}-1,6-naphthyridin-2(1H)-one, Glutathione S-transferase class-mu 26 kDa isozyme,Serine/threonine-protein kinase PAK 1 | | Authors: | Wang, W, Oh, A, Kiefer, J.R, Hsu, P.L. | | Deposit date: | 2025-02-03 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Integrating Hydrogen Exchange with Molecular Dynamics for Improved Ligand Binding Predictions.
J.Chem.Inf.Model., 65, 2025
|
|
5LIU
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
9N48
 
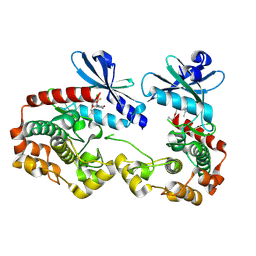 | | Crystal structure of PAK1 bound to compound C1 | | Descriptor: | (6M)-8-[2-(2-aminoethoxy)ethyl]-6-[2-chloro-3-fluoro-4-(2-oxopyrrolidin-1-yl)phenyl]-2-(ethylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Glutathione S-transferase class-mu 26 kDa isozyme,Serine/threonine-protein kinase PAK 1 | | Authors: | Wang, W, Oh, A, Kiefer, J.R, Hsu, P.L. | | Deposit date: | 2025-02-02 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Integrating Hydrogen Exchange with Molecular Dynamics for Improved Ligand Binding Predictions.
J.Chem.Inf.Model., 65, 2025
|
|
8SWT
 
 | |
5E44
 
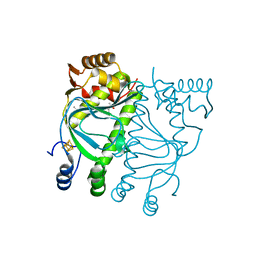 | | Crystal structure of holo-FNR of A. fischeri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FNR regulator, IRON/SULFUR CLUSTER | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium.
Sci Adv, 1, 2015
|
|
8YSV
 
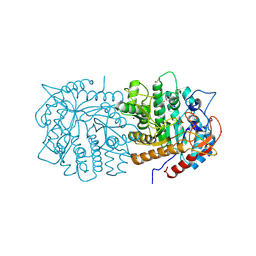 | | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase, ETHANOL, ... | | Authors: | Wang, W.Y, Li, Y.J, Liu, Z.Y, Han, X.D. | | Deposit date: | 2024-03-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis
To Be Published
|
|
8SL9
 
 | | Crystal structure of Francisella tularensis HPPK-DHPS in complex with HPPK inhibitor HP-73 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]-5'-thioadenosine, ... | | Authors: | Shaw, G.X, Shi, G, Cherry, S, Needle, D, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Francisella tularensis HPPK-DHPS in complex with HPPK inhibitor HP-73
To be published
|
|
