4OJD
 
 | |
7PMU
 
 | | Crystal structure of native Iripin-8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, Serpin-8, ... | | Authors: | Polderdijk, S, Kotal, J, Chmelar, J, Huntington, J.A. | | Deposit date: | 2021-09-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Ixodes ricinus Salivary Serpin Iripin-8 Inhibits the Intrinsic Pathway of Coagulation and Complement.
Int J Mol Sci, 22, 2021
|
|
7PQ3
 
 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in complex with its post-catalytic reaction product | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ2
 
 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in its apo form | | Descriptor: | CRISPR-associated protein, APE2256 family, CRISPR Ring Nuclease | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ6
 
 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12A from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQA
 
 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12G/K169G from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PFJ
 
 | |
7PF8
 
 | | SynFtn Variant E141A | | Descriptor: | CHLORIDE ION, Ferritin, SODIUM ION | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
7PFB
 
 | |
7PFI
 
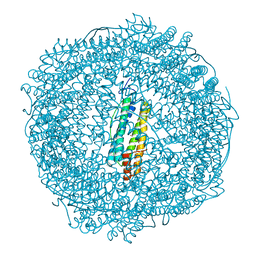 | |
7PF7
 
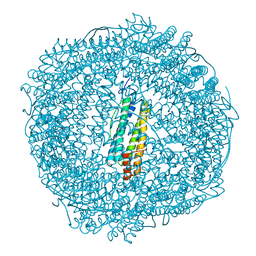 | | Apo structure of SynFtn variant D65A | | Descriptor: | CHLORIDE ION, Ferritin, SODIUM ION | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
7PFK
 
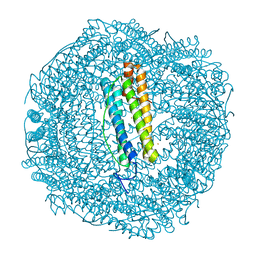 | |
7PFG
 
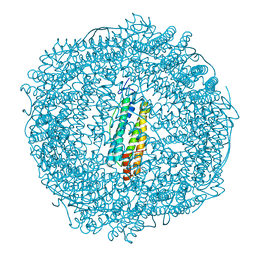 | |
7PFH
 
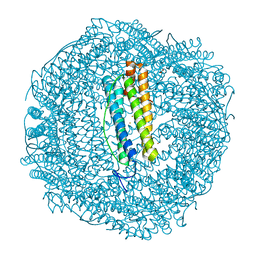 | | 2 minute Fe2+ soak structure of SynFtn E141D | | Descriptor: | ACETATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
6RQS
 
 | | RW16 peptide | | Descriptor: | ARG-ARG-TRP-ARG-ARG-TRP-TRP-ARG-ARG-TRP-TRP-ARG-ARG-TRP-ARG-ARG | | Authors: | Jobin, M.-L, Grelard, A, Alves, I, Mackereth, C.D. | | Deposit date: | 2019-05-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Biophysical Insight on the Membrane Insertion of an Arginine-Rich Cell-Penetrating Peptide.
Int J Mol Sci, 20, 2019
|
|
1N9S
 
 | | Crystal structure of yeast SmF in spacegroup P43212 | | Descriptor: | Small nuclear ribonucleoprotein F | | Authors: | Collins, B.M, Cubeddu, L, Naidoo, N, Harrop, S.J, Kornfeld, G.D, Dawes, I.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2002-11-26 | | Release date: | 2002-12-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Homomeric ring assemblies of eukaryotic Sm proteins have affinity for
both RNA and DNA: Crystal structure of an oligomeric complex of yeast
SmF
J.Biol.Chem., 278, 2003
|
|
5N65
 
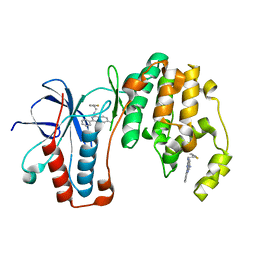 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9h | | Descriptor: | 2-phenyl-~{N}4-(2-thiophen-2-ylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N64
 
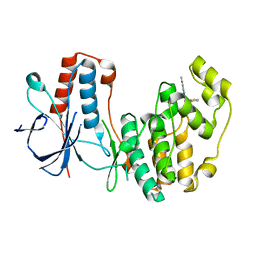 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9g | | Descriptor: | 2-phenyl-~{N}4-(thiophen-2-ylmethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
7Q4R
 
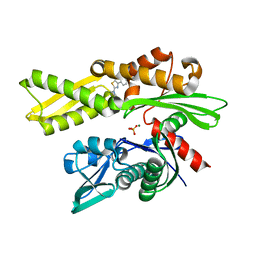 | | Crystal structure of human HSP72-NBD in complex with fragment 1 | | Descriptor: | 4-(4-phenyl-1,3-thiazol-2-yl)piperazine-1-carboxamide, Heat shock 70 kDa protein 1A, MAGNESIUM ION, ... | | Authors: | Le Bihan, Y.V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and Characterization of a Cryptic Secondary Binding Site in the Molecular Chaperone HSP70.
Molecules, 27, 2022
|
|
1E0C
 
 | | SULFURTRANSFERASE FROM AZOTOBACTER VINELANDII | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bordo, D, Deriu, D, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-08 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of a Sulfurtransferase from Azotobacter Vinelandii Highlights the Evolutionary Relationship between the Rhodanese and Phosphatase Enzyme Families
J.Mol.Biol., 298, 2000
|
|
1ZYZ
 
 | | Structures of Yeast Ribonucloetide Reductase I | | Descriptor: | GLYCINE, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5N66
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9j | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 14, ~{N}4-[[4-(cyclopropylmethyl)furan-2-yl]methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
2BW3
 
 | | Three-dimensional structure of the Hermes DNA transposase | | Descriptor: | TRANSPOSASE | | Authors: | Hickman, A.B, Perez, Z.N, Zhou, L, Musingarimi, P, Ghirlando, R, Hinshaw, J.E, Craig, N.L, Dyda, F. | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of a Eukaryotic DNA Transposase
Nat.Struct.Mol.Biol., 12, 2005
|
|
5FDY
 
 | |
5FLK
 
 | |
