5MKY
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH 4-chloro-2-methyl-5-((2-methyl-1,2,3,4-tetrahydroisoquinolin-5-yl)amino)pyridazin-3(2H)-one | | Descriptor: | 4-chloranyl-2-methyl-5-[(2-methyl-3,4-dihydro-1~{H}-isoquinolin-5-yl)amino]pyridazin-3-one, Bromodomain-containing protein 9 | | Authors: | Chung, C.-W. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of a Potent, Cell Penetrant, and Selective p300/CBP-Associated Factor (PCAF)/General Control Nonderepressible 5 (GCN5) Bromodomain Chemical Probe.
J. Med. Chem., 60, 2017
|
|
9LIA
 
 | | Crystal structure of AaHPPD-ZD001 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-(1-methyl-5-oxidanyl-pyrazol-4-yl)carbonyl-2-(phenylmethyl)isoindole-1,3-dione, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2025-01-14 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Repurposing 4-Hydroxyphenylpyruvate dioxygenase inhibitors as novel agents for mosquito control: A structure-based design approach.
Int.J.Biol.Macromol., 315, 2025
|
|
6QCG
 
 | | PCNA complex with Cdt1 N-terminal PIP-box peptide | | Descriptor: | DNA replication factor Cdt1, Proliferating cell nuclear antigen | | Authors: | Perrakis, A, von Castelmur, E. | | Deposit date: | 2018-12-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2E3 ligase activity to Cdt1.
Life Sci Alliance, 1, 2018
|
|
8KBM
 
 | | Structure of AcrIIA7 complexed with cGG | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
7MM4
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-115 | | Descriptor: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2021-04-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
8UBR
 
 | | Complex of the phosphorylated human cystic fibrosis transmembrane conductance regulator (CFTR) with CFTRinh-172 and ATP/Mg | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 4-[(Z)-{(3M)-4-oxo-2-sulfanylidene-3-[3-(trifluoromethyl)phenyl]-1,3-thiazolidin-5-ylidene}methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Young, P.G, Fiedorczuk, K, Chen, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for CFTR inhibition by CFTR inh -172.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QNJ
 
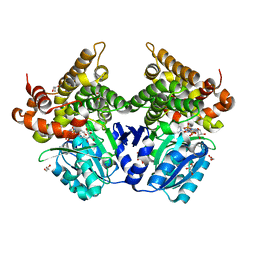 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT F254I COMPLEXED WITH FE, NAD+, AND GLYCEROL | | Descriptor: | FE (III) ION, GLYCEROL, Lactaldehyde reductase, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
8IDX
 
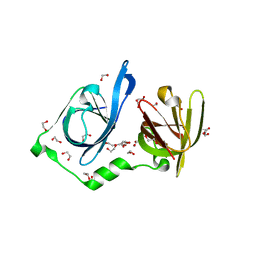 | | Structure of p205 HIN | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Interferon-activable protein 205-B, ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of p205 HIN
to Be published
|
|
6HIA
 
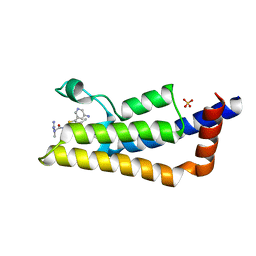 | | The ATAD2 bromodomain in complex with compound 13 | | Descriptor: | (2~{R})-~{N}-[5-(5-azanylpyridin-3-yl)-4-ethanoyl-1,3-thiazol-2-yl]-2-carbamimidamido-propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
8WJD
 
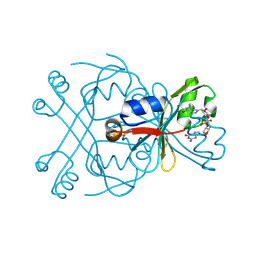 | | Structure of SptTad2 complexed with cGG | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SptTad2 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-25 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
6HI3
 
 | | The ATAD2 bromodomain in complex with compound 4 | | Descriptor: | 2-azanyl-~{N}-(4-ethanoyl-1,3-thiazol-2-yl)-2-methyl-propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
6YJZ
 
 | |
8B0E
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VB158 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-3,4,6-tris(oxidanyl)-2-[2-[2,2,2-tris(fluoranyl)ethanoylamino]ethoxy]cyclohexane-1-carboxylic acid, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
8FH5
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, Aldo-keto reductase family 1 member B1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001
To Be Published
|
|
8FH6
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001
To Be Published
|
|
8B0D
 
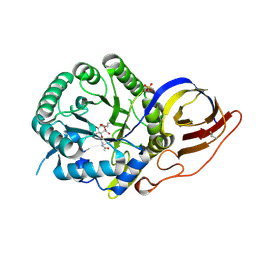 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VB151 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-2-(2-acetamidoethoxy)-3,4,6-tris(oxidanyl)cyclohexane-1-carboxylic acid, ALANINE, SULFATE ION, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
6QV7
 
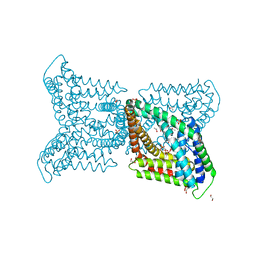 | |
8OOL
 
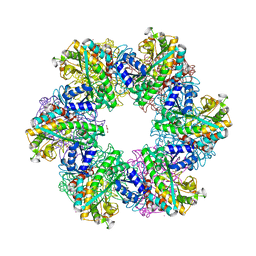 | |
6Z72
 
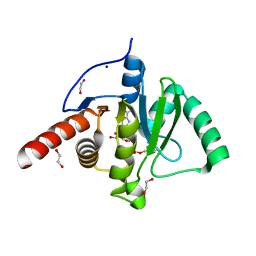 | | SARS-CoV-2 Macrodomain in complex with ADP-HPM | | Descriptor: | 1,2-ETHANEDIOL, Adenosine Diphosphate (Hydroxymethyl)pyrrolidine monoalcohol, D-MALATE, ... | | Authors: | Zorzini, V, Rack, J, Ahel, I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential.
Open Biology, 10, 2020
|
|
8J69
 
 | |
6WFV
 
 | | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Guo, H.-F, Tsai, C.-L, Miller, M.D, Phillips Jr, G.N, Tainer, J.A, Kurie, J.M. | | Deposit date: | 2020-04-04 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a collagen galactosylhydroxylysyl glucosyltransferase from human
To Be Published
|
|
8D7M
 
 | | Human Casein kinase 1 delta in complex with phosphorylated human PERIOD2 FASP peptide | | Descriptor: | Casein kinase I isoform delta, Period circadian protein homolog 2 peptide | | Authors: | Philpott, J.M, Freeberg, A.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Mol.Cell, 83, 2023
|
|
8D7O
 
 | | Human Casein kinase 1 delta in complex with phosphorylated human PERIOD2 FASP peptide | | Descriptor: | Casein kinase I isoform delta, Period circadian protein homolog 2 peptide | | Authors: | Philpott, J.M, Freeberg, A.M, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2022-06-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Mol.Cell, 83, 2023
|
|
5K8J
 
 | | Structure of Caulobacter crescentus VapBC1 (apo form) | | Descriptor: | GLYCEROL, Ribonuclease VapC, VapB family protein | | Authors: | Bendtsen, K.L, Xu, K, Luckmann, M, Brodersen, D.E. | | Deposit date: | 2016-05-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding.
Nucleic Acids Res., 45, 2017
|
|
4ZM7
 
 | |
