3V50
 
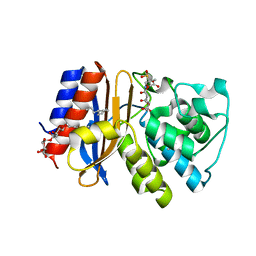 | | Complex of SHV S130G mutant beta-lactamase complexed to SA2-13 | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, Beta-lactamase, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Wei, K, van den Akker, F. | | Deposit date: | 2011-12-15 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The importance of the trans-enamine intermediate as a beta-lactamase inhibition strategy probed in inhibitor-resistant SHV beta-lactamase variants.
Chemmedchem, 7, 2012
|
|
6G3J
 
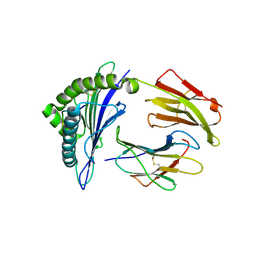 | | MHC A02 Allele presenting MTSAIGILVP | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Rizkallah, P.J, Sewell, A.K. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide Super-Agonist Enhances T-Cell Responses to Melanoma.
Front Immunol, 10, 2019
|
|
3UZJ
 
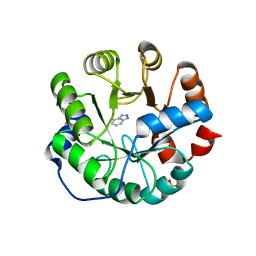 | | Designed protein KE59 R13 3/11H with benzotriazole | | Descriptor: | 1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UPR
 
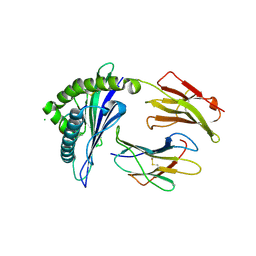 | | HLA-B*57:01 complexed to pep-V and Abacavir | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Pompeu, Y.A, Ostrov, D.A. | | Deposit date: | 2011-11-18 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY8
 
 | | Designed protein KE59 R5_11/5F | | Descriptor: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3FA3
 
 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, trigonal crystal form | | Descriptor: | 2,2-difluoro-3,3-dihydroxybutanedioic acid, 2,3-dimethylmalate lyase, GLYCEROL, ... | | Authors: | Narayanan, B.C, Herzberg, O. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
3UTH
 
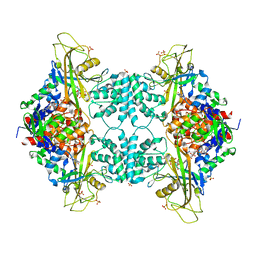 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase complexed with substrate UDP-Galp in reduced state | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
3F7K
 
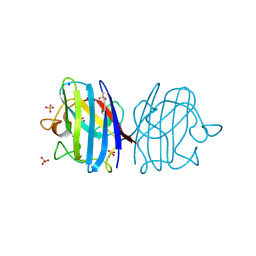 | | X-ray Crystal Structure of an Alvinella pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper,Zinc Superoxide Dismutase, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
3FC1
 
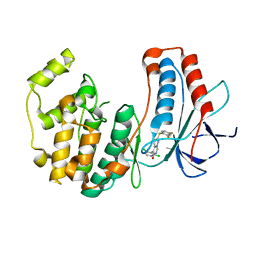 | | Crystal structure of p38 kinase bound to pyrimido-pyridazinone inhibitor | | Descriptor: | 5-(2,6-dichlorophenyl)-2-[(2,4-difluorophenyl)sulfanyl]-6H-pyrimido[1,6-b]pyridazin-6-one, CHLORIDE ION, Mitogen-activated protein kinase 14 | | Authors: | Jacobs, M.D, Bellon, S. | | Deposit date: | 2008-11-20 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evaluating the molecular mechanics poisson-boltzmann surface area free energy method using a congeneric series of ligands to p38 MAP kinase.
J.Med.Chem., 48, 2005
|
|
3V5M
 
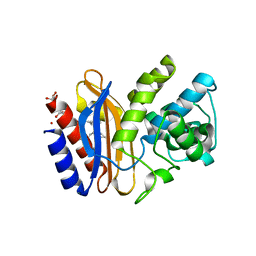 | |
3VBL
 
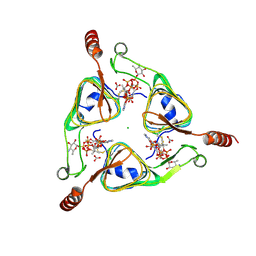 | | Crystal Structure of the S84C mutant of AntD, an N-acyltransferase from Bacillus cereus in complex with dTDP-4-amino-4,6-dideoxyglucose and Coenzyme A | | Descriptor: | BICARBONATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kubiak, R.L, Holden, H.M. | | Deposit date: | 2012-01-02 | | Release date: | 2012-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of AntD: An N-Acyltransferase Involved in the Biosynthesis of d-Anthrose.
Biochemistry, 51, 2012
|
|
3V9I
 
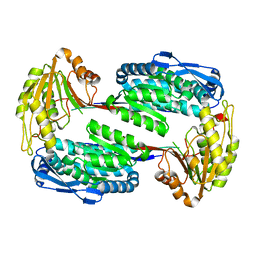 | |
3VBN
 
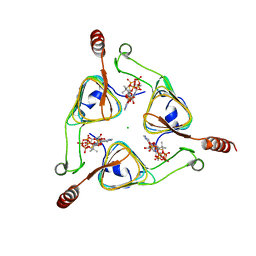 | |
3DW5
 
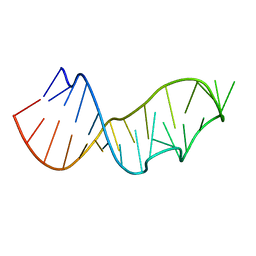 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23S rRNA, U2656-OCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3VBK
 
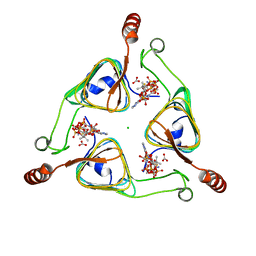 | | Crystal Structure of the S84A mutant of AntD, an N-acyltransferase from Bacillus cereus in complex with dTDP-4-amino-4,6-dideoxyglucose and Coenzyme A | | Descriptor: | BICARBONATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kubiak, R.L, Holden, H.M. | | Deposit date: | 2012-01-02 | | Release date: | 2012-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies of AntD: An N-Acyltransferase Involved in the Biosynthesis of d-Anthrose.
Biochemistry, 51, 2012
|
|
3DW6
 
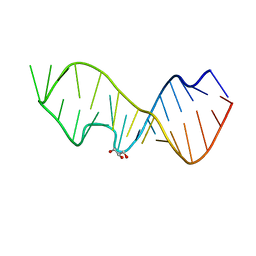 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
4AQN
 
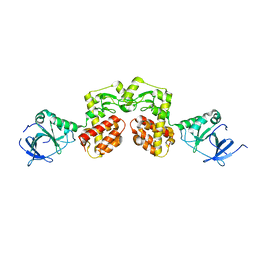 | | Crystal structure of pesticin from Y. pestis | | Descriptor: | MAGNESIUM ION, PESTICIN | | Authors: | Zeth, K, Albrecht, R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
3E1I
 
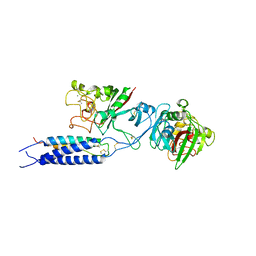 | |
3GAB
 
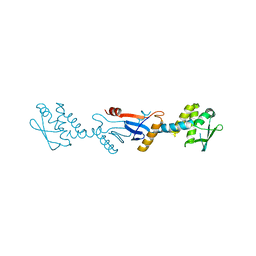 | | C-terminal domain of Bacillus subtilis MutL crystal form I | | Descriptor: | DNA mismatch repair protein mutL | | Authors: | Guarne, A, Pillon, M.C, Lorenowicz, J.J, Mitchell, R.R, Chung, Y.S, Friedhoff, P. | | Deposit date: | 2009-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the endonuclease domain of MutL: unlicensed to cut.
Mol.Cell, 39, 2010
|
|
4AOQ
 
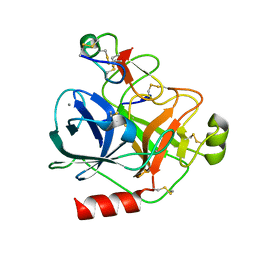 | | Cationic trypsin in complex with mutated Spinacia oleracea trypsin inhibitor III (SOTI-III) (F14A) | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, PENTAETHYLENE GLYCOL, ... | | Authors: | Schmelz, S, Glotzbach, B, Reinwarth, M, Christmann, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Spinacia Oleracea Trypsin Inhibitor III (Soti-III)
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3V9K
 
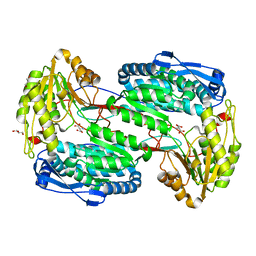 | |
3V9L
 
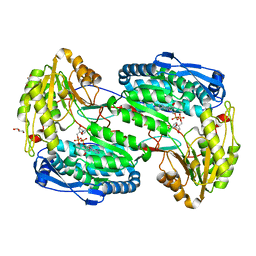 | |
3VBI
 
 | | Crystal Structure of AntD, an N-acyltransferase from Bacillus cereus in complex with dTDP-4-amino-4,6-dideoxyglucose and Coenzyme A | | Descriptor: | BICARBONATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kubiak, R.L, Holden, H.M. | | Deposit date: | 2012-01-02 | | Release date: | 2012-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of AntD: An N-Acyltransferase Involved in the Biosynthesis of d-Anthrose.
Biochemistry, 51, 2012
|
|
3FA4
 
 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, triclinic crystal form | | Descriptor: | 2,3-dimethylmalate lyase, MAGNESIUM ION | | Authors: | Narayanan, B.C, Herzberg, O. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
4AOR
 
 | | Cationic trypsin in complex with the Spinacia oleracea trypsin inhibitor III (SOTI-III) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Schmelz, S, Glotzbach, B, Reinwarth, M, Christmann, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural Characterization of Spinacia Oleracea Trypsin Inhibitor III (Soti-III)
Acta Crystallogr.,Sect.D, 69, 2013
|
|
