2FL1
 
 | | Crystal structure of red fluorescent protein from Zoanthus, zRFP574, at 2.4A resolution | | Descriptor: | Red fluorescent protein zoanRFP, SULFATE ION | | Authors: | Pletnev, V, Pletneva, N, Martynov, V, Tikhonova, T, Popov, B, Pletnev, S. | | Deposit date: | 2006-01-05 | | Release date: | 2007-01-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a red fluorescent protein from Zoanthus, zRFP574, reveals a novel chromophore
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2FM2
 
 | | HCV NS3-4A protease domain complexed with a ketoamide inhibitor, SCH446211 | | Descriptor: | BETA-MERCAPTOETHANOL, NS3 protease/helicase, NS4a protein, ... | | Authors: | Yi, M, Tong, X, Skelton, A, Chase, R, Chen, T, Prongay, A, Bogen, S.L, Saksena, A.K, Njoroge, F.G, Veselenak, R.L, Pyles, R.B, Bourne, N, Malcolm, B.A, Lemon, S.M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations
J.Biol.Chem., 281, 2006
|
|
2FI7
 
 | |
2I2X
 
 | | Crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOB(III)AMIDE, Methyltransferase 1, POTASSIUM ION, ... | | Authors: | Hagemeier, C.H, Kruer, M, Thauer, R.K, Warkentin, E, Ermler, U. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GSY
 
 | | The 2.6A structure of Infectious Bursal Virus Derived T=1 Particles | | Descriptor: | CALCIUM ION, polyprotein | | Authors: | Garriga, D, Querol-Audi, J, Abaitua, F, Saugar, I, Pous, J, Verdaguer, N, Caston, J.R, Rodriguez, J.F. | | Deposit date: | 2006-04-27 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6-angstrom structure of infectious bursal disease virus-derived t=1 particles reveals new stabilizing elements of the virus capsid.
J.Virol., 80, 2006
|
|
2GQM
 
 | | Solution structure of Human Cu(I)-Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GWE
 
 | |
2IWV
 
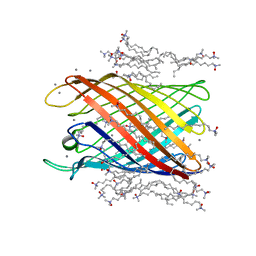 | | Structure of the monomeric outer membrane porin OmpG in the open and closed conformation | | Descriptor: | CALCIUM ION, LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE PROTEIN G, ... | | Authors: | Yildiz, O, Vinothkumar, K.R, Goswami, P, Kuehlbrandt, W. | | Deposit date: | 2006-07-04 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Monomeric Outer-Membrane Porin Ompg in the Open and Closed Conformation.
Embo J., 25, 2006
|
|
2IV1
 
 | |
2H16
 
 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
2GQL
 
 | | Solution structure of Human Ni(II)-Sco1 | | Descriptor: | NICKEL (II) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2IHQ
 
 | |
2J3Z
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 6.1 | | Descriptor: | C2 TOXIN COMPONENT I, COBALT (II) ION, GLYCEROL, ... | | Authors: | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | Deposit date: | 2006-08-23 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
2IKQ
 
 | |
2IUO
 
 | | Site Directed Mutagenesis of Key Residues Involved in the Catalytic Mechanism of Cyanase | | Descriptor: | AZIDE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, M.P. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
2J7A
 
 | | Crystal structure of cytochrome c nitrite reductase NrfHA complex from Desulfovibrio vulgaris | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE NRFA, ... | | Authors: | Rodrigues, M.L, Oliveira, T.F, Pereira, I.A.C, Archer, M. | | Deposit date: | 2006-10-06 | | Release date: | 2006-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure of the Membrane-Bound Cytochrome C Quinol Dehydrogenase Nrfh Reveals Novel Haem Coordination.
Embo J., 25, 2006
|
|
2J28
 
 | | MODEL OF E. COLI SRP BOUND TO 70S RNCS | | Descriptor: | 23S RIBOSOMAL RNA, 4.5S SIGNAL RECOGNITION PARTICLE RNA, 50S ribosomal protein L11, ... | | Authors: | Halic, M, Blau, M, Becker, T, Mielke, T, Pool, M.R, Wild, K, Sinning, I, Beckmann, R. | | Deposit date: | 2006-08-16 | | Release date: | 2006-11-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Following the Signal Sequence from Ribosomal Tunnel Exit to Signal Recognition Particle
Nature, 444, 2006
|
|
2J9F
 
 | | Human branched-chain alpha-ketoacid dehydrogenase-decarboxylase E1b | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Jun, L, Machius, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2006-11-07 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Two Active Sites in Human Branched-Chain Alpha- Keto Acid Dehydrogenase Operate Independently without an Obligatory Alternating-Site Mechanism.
J.Biol.Chem., 282, 2007
|
|
2ICZ
 
 | | NMR Structures of the Expanded DNA 10bp xTGxTAxCxGCxAxGT:xACTxGCGxTAxCA | | Descriptor: | 5'-D(*(XAE)P*CP*TP*(XGA)P*CP*GP*(XTY)P*AP*(XCS)P*A)-3', 5'-D(*(XTY)P*GP*(XTY)P*AP*(XCS)P*(XGA)P*CP*(XAE)P*(XGA)P*T)-3' | | Authors: | Lynch, S.R. | | Deposit date: | 2006-09-13 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Toward a Designed, Functioning Genetic System with Expanded-Size Base Pairs: Solution Structure of the Eight-Base xDNA Double Helix.
J.Am.Chem.Soc., 128, 2006
|
|
2J58
 
 | | The structure of Wza | | Descriptor: | HEXANE, N-OCTANE, OUTER MEMBRANE LIPOPROTEIN WZA, ... | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-09-12 | | Release date: | 2006-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Wza the Translocon for E. Coli Capsular Polysaccharides Defines a New Class of Membrane Protein.
Nature, 444, 2006
|
|
6QDF
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000096 | | Descriptor: | 3-[[6-tert-butyl-2-[methyl-[(3S)-1-methylpyrrolidin-3-yl]amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QKY
 
 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6QDE
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000877 | | Descriptor: | 3-[[6-~{tert}-butyl-2-[3-(dimethylamino)propyl-methyl-amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
8B9P
 
 | | ACE2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-GLY-ARG-GLN-PHE-CYS-HIS-THR-LEU-MET-PRO-ARG-HIS-LEU-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
6K1N
 
 | | PLP-bound form of a putative cystathionine gamma-lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, S, Wang, Y. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural characterization of cystathionine gamma-lyase smCSE enables aqueous metal quantum dot biosynthesis.
Int.J.Biol.Macromol., 174, 2021
|
|
