7O6Z
 
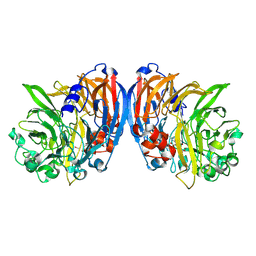 | | Structure of a neodymium-containing, XoxF1-type methanol dehydrogenase | | Descriptor: | METHANOL, Methanol dehydrogenase (Cytochrome c) subunit 1, Neodymium Ion, ... | | Authors: | Schmitz, R, Dietl, A, Op den Camp, H, Barends, T. | | Deposit date: | 2021-04-12 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Neodymium as Metal Cofactor for Biological Methanol Oxidation: Structure and Kinetics of an XoxF1-Type Methanol Dehydrogenase.
Mbio, 12, 2021
|
|
5LPP
 
 | | tRNA guanine Transglycosylase (TGT) in co-crystallized complex (space group C2) with 6-amino-4-(2-((3aR,4R,6R,6aR)-6-methoxy-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)ethyl)-2-(methylamino)-1H-imidazo[4,5-g]quinazolin-8(7H)-one | | Descriptor: | 4-[2-[(3~{a}~{R},4~{R},6~{R},6~{a}~{R})-4-methoxy-2,2-dimethyl-3~{a},4,6,6~{a}-tetrahydrofuro[3,4-d][1,3]dioxol-6-yl]ethyl]-6-azanyl-2-(methylamino)-1,7-dihydroimidazo[4,5-g]quinazolin-8-one, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2016-08-14 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Carbohydrate-based Inhibitors targeting the Ribose-34 pocket of Z.mobilis TGT and changing the oligomeric state of the homodimer
To be published
|
|
4W5K
 
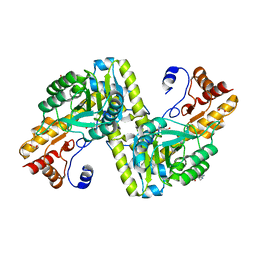 | |
5LIU
 
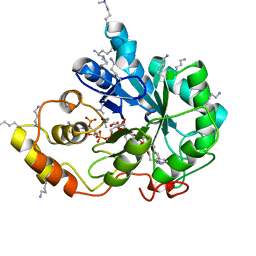 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor IDD388 | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | Deposit date: | 2016-07-15 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LXI
 
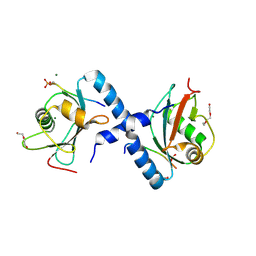 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | 1,2-ETHANEDIOL, Cysteine protease ATG4B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
7OKW
 
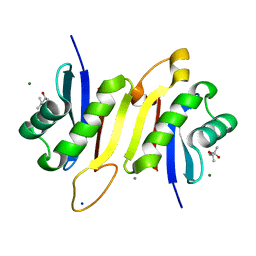 | | 1.62A X-ray crystal structure of the conserved C-terminal (CCT) of human OSR1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Bax, B.D, Elvers, K.T, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
7O86
 
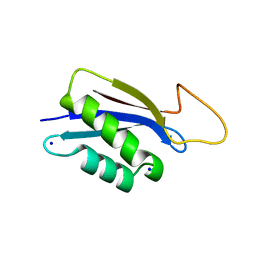 | | 1.73A X-ray crystal structure of the conserved C-terminal (CCT) of human SPAK | | Descriptor: | CALCIUM ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Elvers, K.T, Bax, B.D, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-04-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
4UZS
 
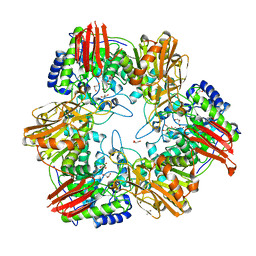 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpah-Galactose.
FEBS J., 283, 2016
|
|
8GOA
 
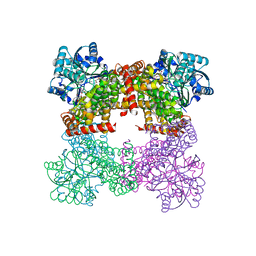 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
6P0Q
 
 | | Crystal Structure of Ubiquitin-like Domain of Human WDR12 | | Descriptor: | 1,2-ETHANEDIOL, Ribosome biogenesis protein WDR12 | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-17 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of Ubiquitin-like Domain of Human WDR12
to be published
|
|
8PXZ
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with natural substrate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-ALANINE, ... | | Authors: | de Munnik, M, Schofield, C.J. | | Deposit date: | 2023-07-24 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Biochemical and crystallographic studies of L,D-transpeptidase 2 from Mycobacterium tuberculosis with its natural monomer substrate.
Commun Biol, 7, 2024
|
|
6PKI
 
 | | Zebrafish N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA) catalytic domain (C56S C230S) in complex with N-acetyl-alpha-D-glucosamine (alpha-GlcNAc) and mannose 6-phosphate (M6P) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-alpha-D-mannopyranose, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crystal Structure of the Mannose-6-Phosphate Uncovering Enzyme.
Structure, 28, 2020
|
|
8GOB
 
 | | Crystal Structure of Glycerol Dehydrogenase in the presence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
1JB9
 
 | | Crystal Structure of The Ferredoxin:NADP+ Reductase From Maize Root AT 1.7 Angstroms | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2001-06-03 | | Release date: | 2001-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues.
Biochemistry, 40, 2001
|
|
8Q7O
 
 | | Crystal structure of the FZD3 cysteine-rich domain in complex with a nanobody (14478) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J.S, Zhao, Y, Malinauskas, T, Jones, E.Y. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into Frizzled3 through nanobody modulators.
Nat Commun, 15, 2024
|
|
1IXB
 
 | | CRYSTAL STRUCTURE OF THE E. COLI MANGANESE(II) SUPEROXIDE DISMUTASE MUTANT Y174F AT 0.90 ANGSTROMS RESOLUTION. | | Descriptor: | MANGANESE ION, 1 HYDROXYL COORDINATED, SUPEROXIDE DISMUTASE | | Authors: | Anderson, B.F, Edwards, R.A, Whittaker, M.M, Whittaker, J.W, Baker, E.N, Jameson, G.B. | | Deposit date: | 2002-06-18 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures at 0.90 A resolution of the oxidised and reduced forms of the Y174F mutant of the manganese superoxide dismutase from Escherichia coli
To be Published
|
|
6PI8
 
 | |
4V37
 
 | |
6P42
 
 | |
6PD1
 
 | |
5LWR
 
 | | Endothiapepsin in complex with a derivative of fragment 177 | | Descriptor: | 1,2-ETHANEDIOL, 4-[12-[(1-chloranyl-5,6,7-trimethyl-pyrrolo[3,4-d]pyridazin-3-ium-3-yl)methyl]-10,11-dimethyl-3,4,6,7,11-pentazatricyclo[7.3.0.0^{2,6}]dodeca-1(12),2,4,7,9-pentaen-5-yl]-1,2,5-trimethyl-pyrrole-3-carbaldehyde, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | A False-Positive Screening Hit in Fragment-Based Lead Discovery: Watch out for the Red Herring.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7ORC
 
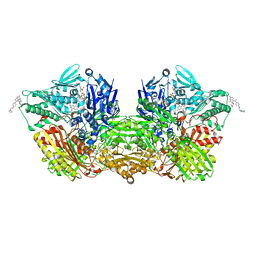 | | Human Aldehyde Oxidase in complex with Raloxifene | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-05 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
6P7O
 
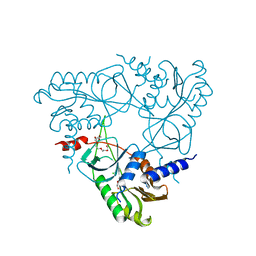 | | Structure of E. coli MS115-1 NucC, Apo form | | Descriptor: | CHLORIDE ION, E. coli MS115-1 NucC, HEXAETHYLENE GLYCOL, ... | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
8GS8
 
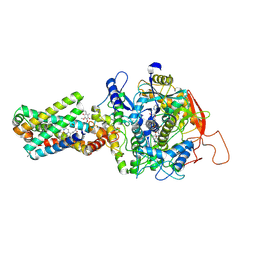 | | cryo-EM structure of the human respiratory complex II | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Du, Z, Zhou, X, Lai, Y, Xu, J, Zhang, Y, Zhou, S, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-09-05 | | Release date: | 2023-05-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the human respiratory complex II.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H2U
 
 | | X-ray Structure of photosystem I-LHCI super complex from Chlamydomonas reinhardtii. | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, H, Kubota-Kawai, H, Misumi, Y, Kurisu, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three structures of PSI-LHCI from Chlamydomonas reinhardtii suggest a resting state re-activated by ferredoxin.
Biochim Biophys Acta Bioenerg, 1864, 2023
|
|
