8E3C
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E38
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E3A
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E2Y
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | Genome polyprotein, VP1, VP2 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8DN2
 
 | |
8DN5
 
 | |
8DN3
 
 | |
8DN4
 
 | | Cryo-EM structure of human Glycine Receptor alpha-1 beta heteromer, glycine-bound state3(desensitized state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-1, Glycine receptor subunit beta,Green fluorescent protein,Glycine receptor beta, ... | | Authors: | Liu, X, Wang, W. | | Deposit date: | 2022-07-10 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric gating of a human hetero-pentameric glycine receptor.
Nat Commun, 14, 2023
|
|
7LZQ
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-4425 | | Descriptor: | B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, N-(3-chloropyridin-4-yl)-2-(5-methyl-4-oxo-4,5-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
8DPV
 
 | |
8DPS
 
 | | The structure of the interleukin 11 signalling complex, truncated gp130 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Metcalfe, R.D, Hanssen, E, Griffin, M.D.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant.
Nat Commun, 14, 2023
|
|
8DPT
 
 | | The structure of the IL-11 signalling complex, with full-length extracellular gp130 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Metcalfe, R.D, Hanssen, E, Griffin, M.D.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant.
Nat Commun, 14, 2023
|
|
8DPW
 
 | | The structure of Interleukin-11 Mutein | | Descriptor: | Interleukin-11, SULFATE ION | | Authors: | Metcalfe, R.D, Griffin, M.D.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant.
Nat Commun, 14, 2023
|
|
6FIK
 
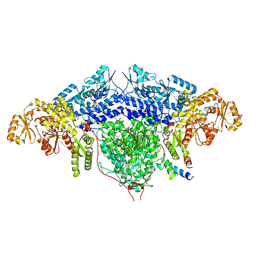 | | ACP2 crosslinked to the KS of the loading/condensing region of the CTB1 PKS | | Descriptor: | Polyketide synthase | | Authors: | Herbst, D.A, Huitt-Roehl, C.R, Jakob, R.P, Townsend, C.A, Maier, T. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | The structural organization of substrate loading in iterative polyketide synthases.
Nat. Chem. Biol., 14, 2018
|
|
6IEY
 
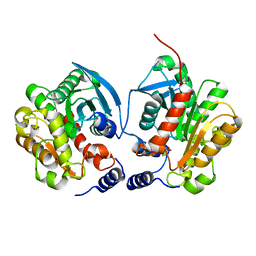 | | Crystal structure of Chloramphenicol-Metabolizaing Enzyme EstDL136-Chloramphenicol complex | | Descriptor: | CHLORAMPHENICOL, Esterase | | Authors: | Kim, S.H, Kang, P.A, Han, K.T, Lee, S.W, Rhee, S.K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome.
PLoS ONE, 14, 2019
|
|
8EH5
 
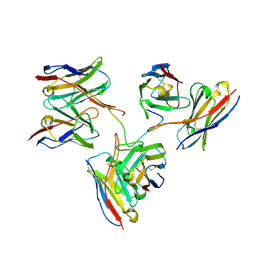 | | Cryo-EM structure of L9 Fab in complex with rsCSP | | Descriptor: | Circumsporozoite protein, L9 Heavy chain, L9 Light chain | | Authors: | Martin, G.M, Ward, A.B. | | Deposit date: | 2022-09-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of epitope selectivity and potent protection from malaria by PfCSP antibody L9.
Nat Commun, 14, 2023
|
|
6FIN
 
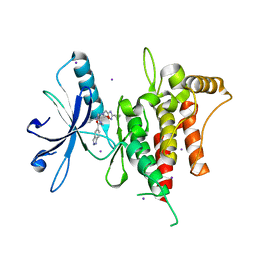 | | DDR1, 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1H-indazole-5-carbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, 1.670A, P1211, Rfree=22.8% | | Descriptor: | 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1~{H}-indazol-5-ylcarbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIQ
 
 | | DDR1, 1-(1H-indazole-5-carbonyl)-5'-methoxy-1'-[2-oxo-2-[(2S)-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, 1.790A, P212121, Rfree=23.8% | | Descriptor: | 1-(1~{H}-indazol-5-ylcarbonyl)-5'-methoxy-1'-[2-oxidanylidene-2-[(2~{S})-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, CHLORIDE ION, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
8E9R
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS in the ligand-free form at 278 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9Q
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant HEX bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9T
 
 | | Crystal structure of wild-type E. coli aspartate aminotransferase in the ligand-free form at 303 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
8E9S
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFCS bound to maleic acid at 278 K | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
6FEW
 
 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.440A, P1211, Rfree=24.1% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
8EKF
 
 | |
8E9V
 
 | | Crystal structure of E. coli aspartate aminotransferase mutant VFIT in the ligand-free form at 303 K | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Chica, R.A, St-Jacques, A.D, Rodriguez, J.M, Thompson, M.C. | | Deposit date: | 2022-08-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Computational remodeling of an enzyme conformational landscape for altered substrate selectivity.
Nat Commun, 14, 2023
|
|
