7ZX6
 
 | | I567L Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZXJ
 
 | | K563A Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZY1
 
 | | I567A Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-23 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.434 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZX5
 
 | | I567T Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.H, Dobbek, H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
7ZXL
 
 | | H93A Mutant of Recombinant CODH-II | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-03-15 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Substrate Activation at the Ni,Fe Cluster of CO Dehydrogenases: The Influence of the Protein Matrix
Acs Catalysis, 2022
|
|
5M10
 
 | |
5Y7T
 
 | | Quaternary complex of AsqJ-Fe3+-2OG-D-cyclopeptin | | Descriptor: | (3R)-4-methyl-3-(phenylmethyl)-1,3-dihydro-1,4-benzodiazepine-2,5-dione, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | Liao, H.J. | | Deposit date: | 2017-08-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the Desaturation of Cyclopeptin and its C3 Epimer Catalyzed by a non-Heme Iron Enzyme: Structural Characterization and Mechanism Elucidation.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8OUC
 
 | | Escherichia coli DPS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA protection during starvation protein | | Authors: | Kovalenko, V.V, Loiko, N.G, Tereshkina, K.B, Tereshkin, E.V, Chulichkov, A.L, Popov, A.N, Krupyanskii, Y.F. | | Deposit date: | 2023-04-22 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Escherichia coli DPS
To be published
|
|
9B17
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PAN1 inhibitor | | Descriptor: | (5P)-5-(1H-indol-3-yl)-1-[2-(piperazin-1-yl)ethyl]-1H-1,2,3-benzotriazole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Geeraerts, Z, Yeh, S.-R. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
6SYM
 
 | |
9GZQ
 
 | | Structure of ForCE lacking the Helical Membrane Plug-in (HMP; DUF1641) | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, ForE1, ... | | Authors: | Arnoux, P, Cherrier, M.V, Nicolet, Y, Legrand, P, Broc, M, Seduk, F, Arias-Cartin, R, Magalon, A, Walburger, A. | | Deposit date: | 2024-10-04 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (3.626 Å) | | Cite: | A scaffold for quinone channeling between membrane and soluble bacterial oxidoreductases.
Nat.Struct.Mol.Biol., 2025
|
|
8BBM
 
 | |
8J1K
 
 | | co-crystal structure of non-carboxylic acid inhibitor with PHD2 | | Descriptor: | Egl nine homolog 1, MANGANESE (II) ION, N-[(6-cyanopyridin-3-yl)methyl]-5-oxidanyl-2-[(3R)-3-oxidanylpyrrolidin-1-yl]-1,7-naphthyridine-6-carboxamide | | Authors: | Xu, J, Fu, Y, Ding, X, Meng, Q, Wang, L, Zhang, M, Ding, X, Ren, F, Zhavoronkov, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | co-crystal structure of non-carboxylic acid inhibitor with PHD2
To Be Published
|
|
5FPB
 
 | |
6JNL
 
 | | REF6 ZnF2-4-NAC004 complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
5FPA
 
 | |
9C0R
 
 | |
6NHG
 
 | | Rhodobacter sphaeroides Mitochondrial respiratory chain complex | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Xia, D, Zhou, F, Esser, L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bacterial cytochromebc1in complex with azoxystrobin reveals a conformational switch of the Rieske iron-sulfur protein subunit.
J.Biol.Chem., 294, 2019
|
|
3KPX
 
 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
6T8M
 
 | | Prolyl Hydroxylase (PHD) involved in hypoxia sensing by Dictyostelium discoideum | | Descriptor: | CHLORIDE ION, GLYCEROL, N-OXALYLGLYCINE, ... | | Authors: | Chowdhury, R, McDonough, M.A, Liu, T, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2019-10-24 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and biophysical analyses of hypoxia sensing prolyl hydroxylases from Dictyostelium discoideum and Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
6HIF
 
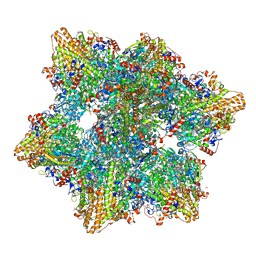 | | Kuenenia stuttgartiensis hydrazine dehydrogenase complex | | Descriptor: | GLYCEROL, HEME C, Hydrazine dehydrogenase, ... | | Authors: | Akram, M, Dietl, A, Mersdorf, U, Prinz, S, Maalcke, W, Keltjens, J, Ferousi, C, de Almeida, N.M, Reimann, J, Kartal, B, Jetten, M.S.M, Parey, K, Barends, T.R.M. | | Deposit date: | 2018-08-29 | | Release date: | 2019-04-17 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A 192-heme electron transfer network in the hydrazine dehydrogenase complex.
Sci Adv, 5, 2019
|
|
5X6R
 
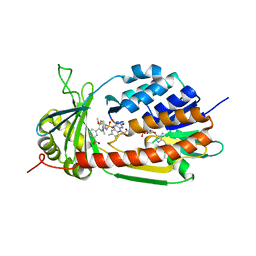 | |
5N7T
 
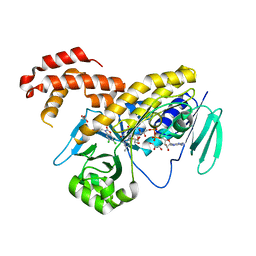 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with 3-(5,6-dichloro-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid | | Descriptor: | 3-(5,6-DICHLORO-2-OXOBENZO[D]OXAZOL-3(2H)-YL)PROPANOIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The discovery of potent and selective kynurenine 3-monooxygenase inhibitors for the treatment of acute pancreatitis.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7C13
 
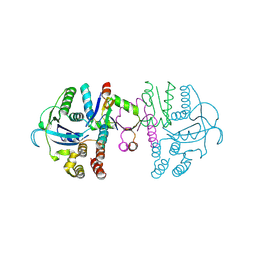 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin, Peptide methionine sulfoxide reductase MsrA | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
7TS6
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 4-methyl-6-(3-((methylamino)methyl)phenyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-{3-[(methylamino)methyl]phenyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-Aminopyridines with a shortened amino sidechain as potent, selective, and highly permeable human neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem., 69, 2022
|
|
