1FQD
 
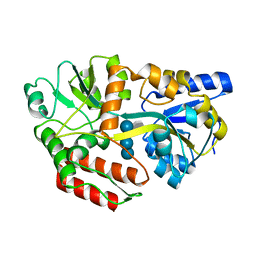 | | CRYSTAL STRUCTURE OF MALTOTETRAITOL BOUND TO CLOSED-FORM MALTODEXTRIN BINDING PROTEIN | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Duan, X, Hall, J.A, Nikaido, H, Quiocho, F.A. | | Deposit date: | 2000-09-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding.
J.Mol.Biol., 306, 2001
|
|
8P4F
 
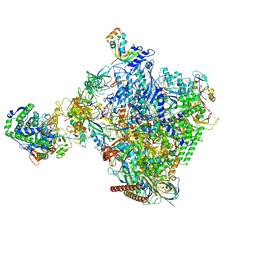 | | Structural insights into human co-transcriptional capping - structure 6 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (38-MER), ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
3EHU
 
 | |
5TTD
 
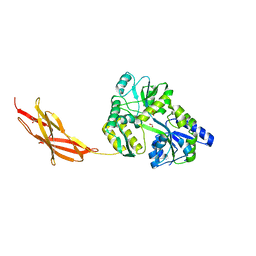 | | Minor pilin FctB from S. pyogenes with engineered intramolecular isopeptide bond | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,Pilin isopeptide linkage domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Young, P.G, Kwon, H, Squire, C.J, Baker, E.N. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Lys-Asn isopeptide bond into an immunoglobulin-like protein domain enhances its stability.
Sci Rep, 7, 2017
|
|
1ERG
 
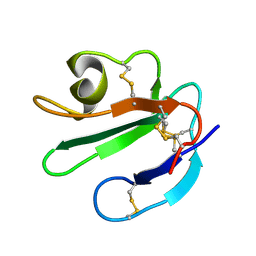 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE EXTRACELLULAR REGION OF THE COMPLEMENT REGULATORY PROTEIN, CD59, A NEW CELL SURFACE PROTEIN DOMAIN RELATED TO NEUROTOXINS | | Descriptor: | CD59 | | Authors: | Kieffer, B, Driscoll, P.C, Campbell, I.D, Willis, A.C, Van Der Merwe, P.A, Davis, S.J. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins.
Biochemistry, 33, 1994
|
|
3H4Z
 
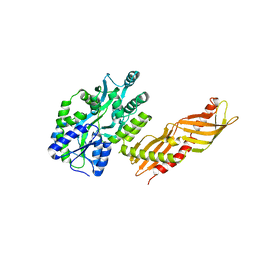 | | Crystal Structure of an MBP-Der p 7 fusion protein | | Descriptor: | Maltose-binding periplasmic protein fused with Allergen DERP7, SODIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pedersen, L.C, Mueller, G.A, London, R.E. | | Deposit date: | 2009-04-21 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins.
J.Allergy Clin.Immunol., 125, 2010
|
|
3H3G
 
 | |
1EZ9
 
 | |
1EZO
 
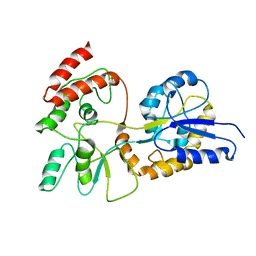 | | GLOBAL FOLD OF MALTODEXTRIN BINDING PROTEIN COMPLEXED WITH BETA-CYCLODEXTRIN | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Mueller, G.A, Choy, W.Y, Yang, D, Forman-Kay, J.D, Venters, R.A, Kay, L.E. | | Deposit date: | 2000-05-11 | | Release date: | 2001-05-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Global folds of proteins with low densities of NOEs using residual dipolar couplings: application to the 370-residue maltodextrin-binding protein.
J.Mol.Biol., 300, 2000
|
|
4CJ0
 
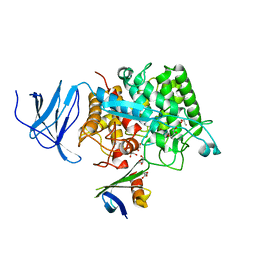 | | Crystal structure of CelD in complex with affitin E12 | | Descriptor: | CALCIUM ION, E12 AFFITIN, ENDOGLUCANASE D, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4CJ1
 
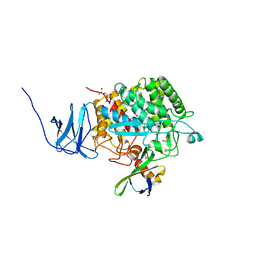 | | Crystal structure of CelD in complex with affitin H3 | | Descriptor: | CALCIUM ION, ENDOGLUCANASE D, GLYCEROL, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4DXB
 
 | |
4EXK
 
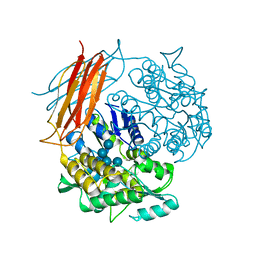 | | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 from Salmonella enterica | | Descriptor: | Maltose-binding periplasmic protein, uncharacterized protein chimera, TRIETHYLENE GLYCOL, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Jedrzejczak, R, Babnigg, G, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-30 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A chimera protein containing MBP fused to the C-terminal domain of the uncharacterized protein STM14_2015 form Salmonella enterica
To be Published
|
|
4FEC
 
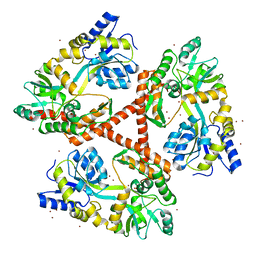 | | Crystal Structure of Htt36Q3H | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4EDQ
 
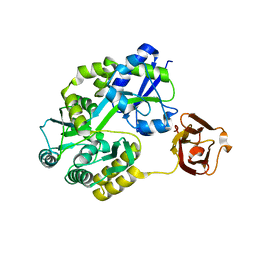 | |
4EGC
 
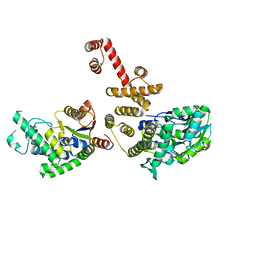 | |
4DXC
 
 | |
4FE8
 
 | | Crystal Structure of Htt36Q3H-EX1-X1-C1(Alpha) | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FED
 
 | | Crystal Structure of Htt36Q3H | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FEB
 
 | | Crystal Structure of Htt36Q3H-EX1-X1-C2(Beta) | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, SODIUM ION, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
6DBV
 
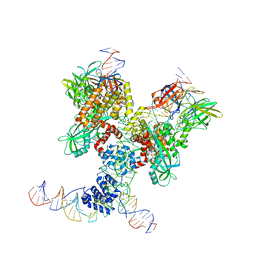 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.291 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D66
 
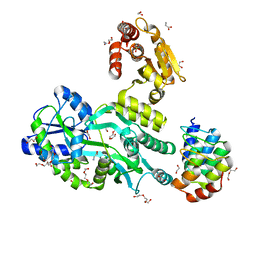 | | Crystal structure of the human dual specificity 1 catalytic domain (C258S) as a maltose binding protein fusion in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, D-ALANINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gumpena, R, Waugh, D.S, Lountos, G.T. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6QYL
 
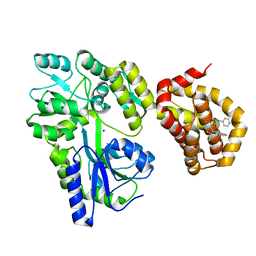 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8CE0
 
 | | N-terminal domain of human apolipoprotein E | | Descriptor: | GLYCEROL, Maltodextrin-binding protein,Apolipoprotein E | | Authors: | Marek, M, Nemergut, M. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
7VQO
 
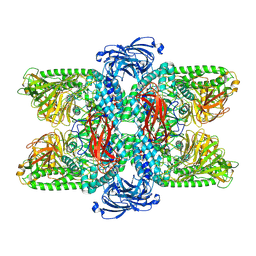 | | Cryo-EM structure of Ams1 bound to the FW domain of Nbr1 | | Descriptor: | Ams1, Nbr1 and malE fusion protein, ZINC ION | | Authors: | Zhang, J, Ye, K. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanism of protein recognition by the FW domain of autophagy receptor Nbr1
Nat Commun, 13, 2022
|
|
