9EOF
 
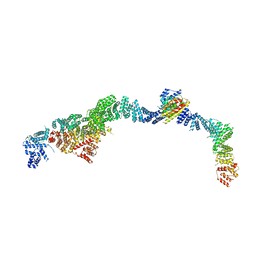 | | Structure of the human INTS5/8/10/15 subcomplex | | Descriptor: | Integrator complex subunit 10, Integrator complex subunit 15, Integrator complex subunit 5, ... | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 84, 2024
|
|
9EOE
 
 | | TF type tau filament from V337M mutant | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Qi, C, Scheres, S.H.W, Michel, G. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Tau filaments with the Alzheimer fold in cases with MAPT mutations V337M and R406W.
Biorxiv, 2024
|
|
9EOC
 
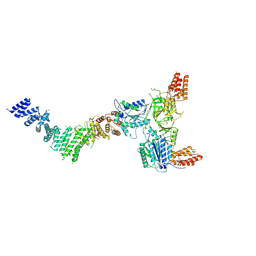 | |
9EO9
 
 | | SF type tau filament from V337M mutant | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Qi, C, Scheres, S.H.W, Michel, G. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tau filaments with the Alzheimer fold in cases with MAPT mutations V337M and R406W.
Biorxiv, 2024
|
|
9EO8
 
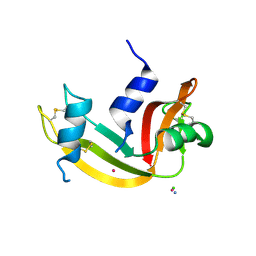 | |
9EO7
 
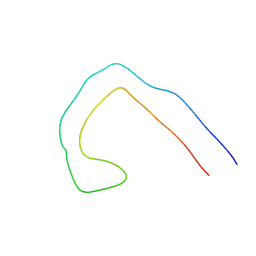 | |
9EO5
 
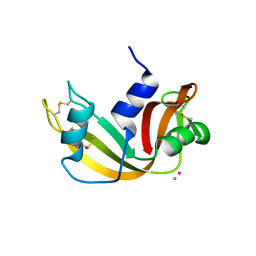 | |
9EO4
 
 | | Outward-open structure of human dopamine transporter bound to cocaine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nielsen, J.C, Salomon, K, Kalenderoglou, I.E, Bargmeyer, S, Pape, T, Shahsavar, A, Loland, C.J. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human dopamine transporter in complex with cocaine
Nature, 2024
|
|
9EO2
 
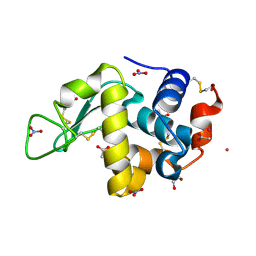 | |
9EO0
 
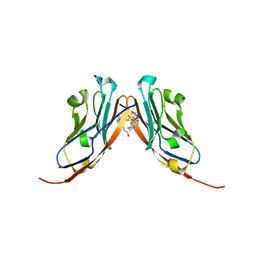 | | Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION, ~{N}-[3-[3-[[5-[(2-hydroxyethylamino)methyl]pyridin-2-yl]carbonylamino]-2-methyl-phenyl]-2-methyl-phenyl]-5-[[3-(methylsulfonylamino)propylamino]methyl]pyridine-2-carboxamide | | Authors: | Plewka, J, Hec, A, Sitar, T, Holak, T. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonsymmetrically Substituted 1,1'-Biphenyl-Based Small Molecule Inhibitors of the PD-1/PD-L1 Interaction.
Acs Med.Chem.Lett., 15, 2024
|
|
9ENZ
 
 | |
9ENT
 
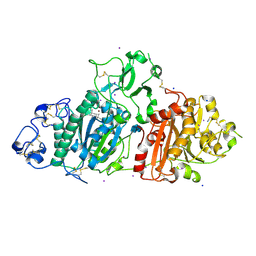 | | SSX structure of Autotaxin in cryogenic conditions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, CALCIUM ION, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Foos, N, Basu, S. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In situ serial crystallography facilitates 96-well plate structural analysis at low symmetry.
Iucrj, 2024
|
|
9ENQ
 
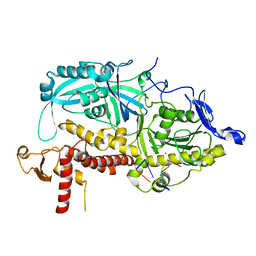 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state active site with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
9ENP
 
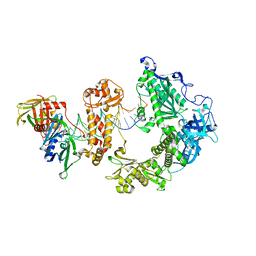 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
9ENF
 
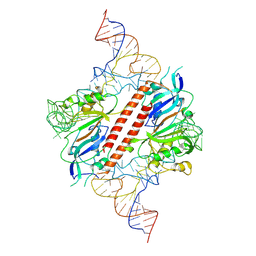 | |
9ENE
 
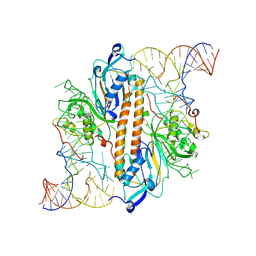 | |
9END
 
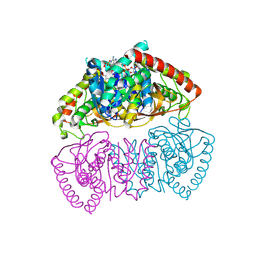 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 3 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Coquille, S, Roche, J, Girard, E, Madern, D. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Navigating the conformational landscape of an enzyme. Stabilization of a low populated conformer by evolutionary mutations triggers Allostery into a non-allosteric enzyme.
To be published
|
|
9ENC
 
 | |
9ENB
 
 | |
9EN6
 
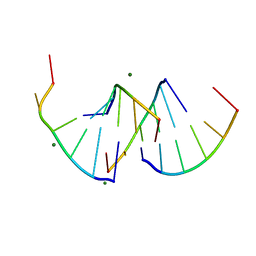 | | Crystal structure of RNA G2C4 repeats - native model pH 6.5 | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Mateja-Pluta, M, Kiliszek, A. | | Deposit date: | 2024-03-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (0.918 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
9EN2
 
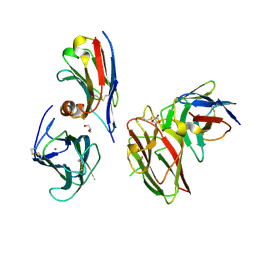 | | Crystal structure of the metalloproteinase enhancer PCPE-1 complexed with nanobodies VHH-H4 and VHH-I5 | | Descriptor: | CALCIUM ION, GLYCEROL, Procollagen C-endopeptidase enhancer 1, ... | | Authors: | Lagoutte, P, Gueguen-Chaignon, V, Bourhis, J.-M, Vadon-Le Goff, S. | | Deposit date: | 2024-03-12 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mono- and Bi-specific Nanobodies Targeting the CUB Domains of PCPE-1 Reduce the Proteolytic Processing of Fibrillar Procollagens.
J.Mol.Biol., 436, 2024
|
|
9EMV
 
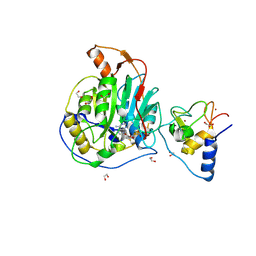 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EML
 
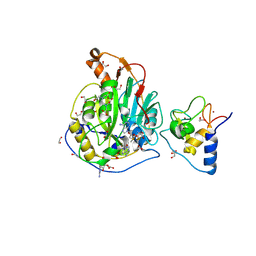 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EMJ
 
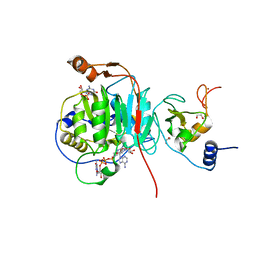 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with Toyocamycin and m7GpppA (Cap0-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EME
 
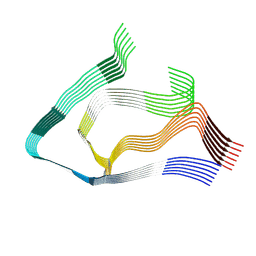 | | AL amyloid fibril from the FOR103 light chain | | Descriptor: | lambda 3 immunoglobulin light chain fragment, residues 2-116 | | Authors: | Pfeiffer, P.B, Karimi-Farsijani, S, Kupfer, N, Schmidt, M, Faendrich, M. | | Deposit date: | 2024-03-08 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Light chain mutations contribute to defining the fibril morphology in systemic AL amyloidosis.
Nat Commun, 15, 2024
|
|
