5O2M
 
 | | p130Cas SH3 domain | | Descriptor: | Breast cancer anti-estrogen resistance 1 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
8DVN
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DVM
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
8DVL
 
 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.18 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
7JQL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-001, mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | Deposit date: | 2020-08-11 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
8ON2
 
 | | NI,FE-CODH -600mV state : 24 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OMY
 
 | | NI,FE-CODH -600mV state : 35 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ZSS
 
 | | Cryo-EM structure of the RO5263397-bound hTAAR1-Gs complex | | Descriptor: | (4~{S})-4-(3-fluoranyl-2-methyl-phenyl)-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
8ZSJ
 
 | | Cryo-EM structure of the apo hTAAR1-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
8II8
 
 | | X-ray crystal structure of pink-colored protein from Pleurotus salmoneostramineus in complex with natural chromophore | | Descriptor: | (2E,5E,7Z,9E,11E,13E,15Z,17E,19Z,21E,23E)-24-methyl-25-oxohexacosa-2,5,7,9,11,13,15,17,19,21,23-undecaenoic acid, ACETYL GROUP, Chromoprotein | | Authors: | Ihara, M, Fukuta, Y, Shirasaka, N. | | Deposit date: | 2023-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the Native Chromoprotein from Pleurotus salmoneostramineus Provides Insights into the Pigmentation Mechanism.
J.Agric.Food Chem., 72, 2024
|
|
6ZDX
 
 | | RIFIN variable region bound to LILRB1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leukocyte immunoglobulin-like receptor subfamily B member 1, ... | | Authors: | Harrison, T.E, Higgins, M.K. | | Deposit date: | 2020-06-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RIFIN-mediated activation of LILRB1 in malaria.
Nature, 587, 2020
|
|
6ZUC
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CsPYL1-Lig1-HAB1 TERNARY COMPLEX | | Descriptor: | 1,4-dimethyl-2-oxidanylidene-~{N}-(phenylmethyl)quinoline-6-sulfonamide, CHLORIDE ION, CSPYL1, ... | | Authors: | Albert, A, Infantes, L, Benavente, J.L. | | Deposit date: | 2020-07-22 | | Release date: | 2021-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided engineering of a receptor-agonist pair for inducible activation of the ABA adaptive response to drought.
Sci Adv, 9, 2023
|
|
8OMX
 
 | | NI,FE-CODH -600mV state : 1 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE(4)-NI(1)-S(5) CLUSTER, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON3
 
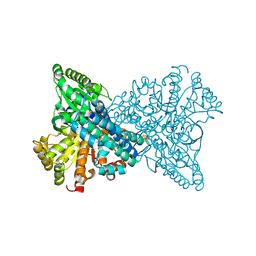 | | NI,FE-CODH -320mV + CN state : 24 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON1
 
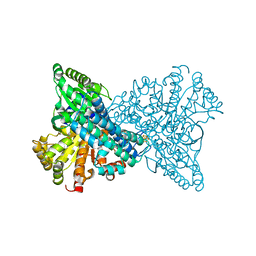 | | NI,FE-CODH -600mV state : 4 h Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ON0
 
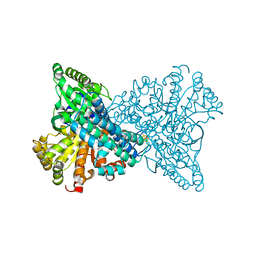 | | NI,FE-CODH -600mV state : 90 min Dioxygen Exposure | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (II) ION, FE (III) ION, ... | | Authors: | Basak, Y, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Stepwise O 2 -Induced Rearrangement and Disassembly of the [NiFe 4 (OH)( mu 3 -S) 4 ] Active Site Cluster of CO Dehydrogenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7JQM
 
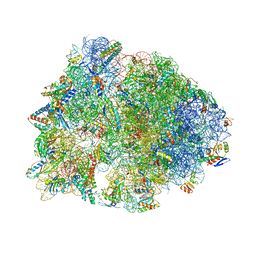 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Bac7-002, mRNA, and deacylated P-site tRNA at 3.05A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mardirossian, M, Sola, R, Beckert, B, Valencic, E, Collis, D.W.P, Borisek, J, Armas, F, Di Stasi, A, Buchmann, J, Syroegin, E.A, Polikanov, Y.S, Magistrato, A, Hilpert, K, Wilson, D.N, Scocchi, M. | | Deposit date: | 2020-08-11 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Peptide Inhibitors of Bacterial Protein Synthesis with Broad Spectrum and SbmA-Independent Bactericidal Activity against Clinical Pathogens.
J.Med.Chem., 63, 2020
|
|
8ZSV
 
 | | Cryo-EM structure of the RO5263397-bound mTAAR1-Gs complex | | Descriptor: | (4~{S})-4-(3-fluoranyl-2-methyl-phenyl)-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
3PRX
 
 | | Structure of Complement C5 in Complex with CVF and SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, ... | | Authors: | Laursen, N.S, Andersen, G.R, Sottrup-Jensen, L, Andersen, K.R, Spillner, E, Braren, I. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex.
Embo J., 30, 2011
|
|
3QCT
 
 | | Crystal structure of the humanized apo LT3015 anti-lysophosphatidic acid antibody Fab fragment | | Descriptor: | LT3015 antibody Fab fragment, heavy chain, light chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1493 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
3QCV
 
 | | Crystal structure of the LT3015 antibody Fab fragment in complex with lysophosphatidic acid (18:2) | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9Z,12Z)-octadeca-9,12-dienoate, LT3015 antibody Fab fragment, heavy chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
3QCU
 
 | | Crystal structure of the LT3015 antibody Fab fragment in complex with lysophosphatidic acid (14:0) | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, LT3015 antibody Fab fragment, heavy chain, ... | | Authors: | Fleming, J.K, Wojciak, J.M, Campbell, M.-A, Huxford, T. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Biochemical and structural characterization of lysophosphatidic Acid binding by a humanized monoclonal antibody.
J.Mol.Biol., 408, 2011
|
|
6BGA
 
 | | 2B4 I-Ek TCR-MHC complex with affinity-enhancing Velcro peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 peptide,MHC I-Ek B chain, ... | | Authors: | Gee, M.H, Sibener, L.V, Birnbaum, M.E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Stress-testing the relationship between T cell receptor/peptide-MHC affinity and cross-reactivity using peptide velcro.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1QF6
 
 | | STRUCTURE OF E. COLI THREONYL-TRNA SYNTHETASE COMPLEXED WITH ITS COGNATE TRNA | | Descriptor: | ADENOSINE MONOPHOSPHATE, THREONINE TRNA, THREONYL-TRNA SYNTHETASE, ... | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 1999-04-06 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site
Cell(Cambridge,Mass.), 97, 1999
|
|
3RJR
 
 | | Crystal Structure of pro-TGF beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1 | | Authors: | Zhu, J.H, Shi, M.L, Springer, T.A. | | Deposit date: | 2011-04-15 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Latent TGF-Beta structure and activation
Nature, 474, 2011
|
|
