2Y5K
 
 | | Orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | 1-[5-(2-METHOXYETHYL)-4-METHYL-THIOPHEN-2-YL]SULFONYL-3-[4-METHOXY-6-(METHYLCARBAMOYLAMINO)PYRIDIN-2-YL]UREA, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Hebeisen, P, Haap, W, Kuhn, B, Mohr, P, Wessel, H.P, Zutter, U, Kirchner, S, Benz, J, Joseph, C, Alvarez-Sanchez, R, Gubler, M, Schott, B, Benardeau, A, Tozzo, E, Kitas, E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3CLK
 
 | |
4KSY
 
 | |
3EE7
 
 | | Crystal Structure of SARS-CoV nsp9 G104E | | Descriptor: | GLYCEROL, PHOSPHATE ION, Replicase polyprotein 1a | | Authors: | Miknis, Z.J, Donaldson, E.F, Umland, T.C, Rimmer, R, Baric, R.S, Schultz, L.W. | | Deposit date: | 2008-09-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth
J.Virol., 83, 2009
|
|
2ZW5
 
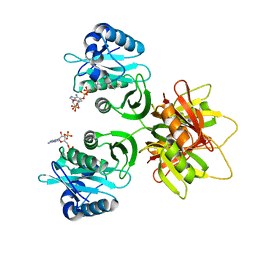 | |
4I9N
 
 | | Crystal structure of rabbit LDHA in complex with AP28161 and AP28122 | | Descriptor: | 6-({2-[(5-chloro-4-{[(2S)-2,3-dihydroxypropyl]oxy}-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, 6-[3-(carboxymethoxy)-5-fluorophenyl]pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4N7P
 
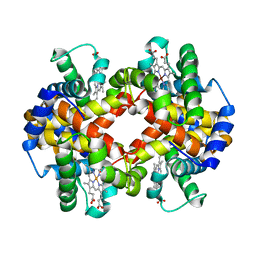 | | Capturing the haemoglobin allosteric transition in a single crystal form; Crystal structure of half-liganded human haemoglobin without phosphate at 2.8 A resolution. | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sugiyama, K, Shibayama, N, Park, S.Y. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Capturing the hemoglobin allosteric transition in a single crystal form
J.Am.Chem.Soc., 136, 2014
|
|
3E2Q
 
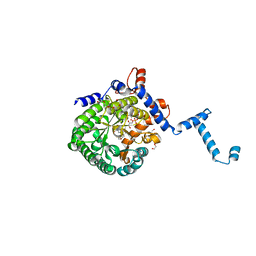 | |
2IY4
 
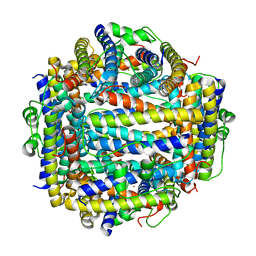 | | X-ray structure of Dps from Listeria monocytogenes | | Descriptor: | FE (III) ION, NON-HEME IRON-CONTAINING FERRITIN | | Authors: | Ilari, A, Bellapadrona, G, Stefanini, S, Chiancone, E. | | Deposit date: | 2006-07-12 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Mutations Lys 114 --> Gln and Asp 126 --> Asn Disrupt an Intersubunit Salt Bridge and Convert Listeria Innocua Dps Into its Natural Mutant Listeria Monocytogenes Dps. Effects on Protein Stability at Low Ph.
Proteins, 66, 2007
|
|
5XPI
 
 | | Structure of UHRF1 TTD in complex with NV01 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, N-[3-(diethylamino)propyl]-2-(12-methyl-9-oxidanylidene-5-thia-1,10,11-triazatricyclo[6.4.0.0^2,6]dodeca-2(6),3,7,11-tetraen-10-yl)ethanamide | | Authors: | Luo, X, Zhao, K. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain
SLAS Discov, 23, 2018
|
|
4D1E
 
 | | THE CRYSTAL STRUCTURE OF HUMAN MUSCLE ALPHA-ACTININ-2 | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Pinotsis, N, Salmazo, A, Sjoeblom, B, Gkougkoulia, E, Djinovic-Carugo, K. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure and Regulation of Human Muscle Alpha-Actinin
Cell(Cambridge,Mass.), 159, 2014
|
|
2J7Y
 
 | | STRUCTURE OF 17-EPIESTRIOL-BOUND ESTROGEN RECEPTOR BETA LBD IN COMPLEX WITH LXXLL MOTIF FROM NCOA5 | | Descriptor: | (16ALPHA,17ALPHA)-ESTRA-1,3,5(10)-TRIENE-3,16,17-TRIOL, 1,2-ETHANEDIOL, BICARBONATE ION, ... | | Authors: | Pike, A.C.W, Brzozowski, A.M, Hubbard, R.E, Walton, J, Bonn, T, Thorsell, A.-G, Engstrom, O, Ljunggren, J, Gustaffson, J.-A, Carlquist, M. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Agonist-Bound Estrogen Receptor Beta Lbd in Complex with Lxxll Motif from Ncoa5
To be Published
|
|
3F1O
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with an internally-bound artificial ligand | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, ... | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4K9R
 
 | | Spore photoproduct lyase Y98F mutant | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, Spore photoproduct lyase, ... | | Authors: | Yang, L, Nelson, R.S, Benjdia, A, Lin, G, Telser, J, Stoll, S, Schlichting, I, Li, L. | | Deposit date: | 2013-04-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A radical transfer pathway in spore photoproduct lyase.
Biochemistry, 52, 2013
|
|
4P43
 
 | |
2ZW7
 
 | |
4P20
 
 | | Crystal structures of the bacterial ribosomal decoding site complexed with amikacin | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Kondo, J, Francois, B, Russell, R.J.M, Murray, J.B, Westhof, E. | | Deposit date: | 2014-02-28 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the gamma-amino-alpha-hydroxybutyryl (haba) group.
Biochimie, 88, 2006
|
|
4KSE
 
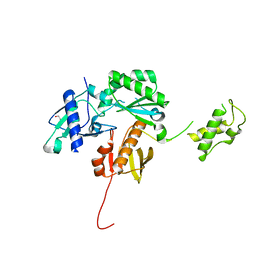 | | Crystal structure of a HIV p51 (219-230) deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, HIV p51 subunit | | Authors: | Zheng, X, Mueller, G.A, Derose, E.F, Pedersen, L.C, Gabel, S.A, Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2013-05-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.
Nucleic Acids Res., 42, 2014
|
|
4NJN
 
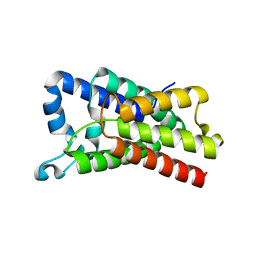 | | Crystal Structure of E.coli GlpG at pH 4.5 | | Descriptor: | Rhomboid protease GlpG | | Authors: | Dickey, S.W, Baker, R.P, Cho, S, Urban, S. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Proteolysis inside the Membrane Is a Rate-Governed Reaction Not Driven by Substrate Affinity.
Cell(Cambridge,Mass.), 155, 2013
|
|
4K1N
 
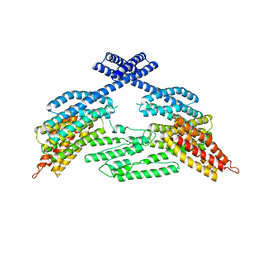 | |
7YHR
 
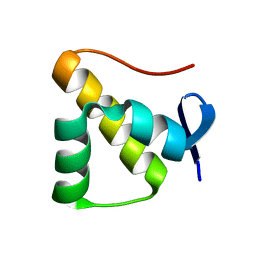 | | Anti-CRISPR protein AcrIC5 | | Descriptor: | Anti-CRISPR protein Type I-C5 | | Authors: | Kang, Y.J, Park, H.H. | | Deposit date: | 2022-07-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structure of the anti-CRISPR protein AcrIC5.
Biochem.Biophys.Res.Commun., 625, 2022
|
|
7XTX
 
 | | High resolution crystal structure of human macrophage migration inhibitory factor in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
7XVX
 
 | |
4I98
 
 | |
4I9U
 
 | | Crystal structure of rabbit LDHA in complex with a fragment inhibitor AP26256 | | Descriptor: | 6-({2-[(5-chloro-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
