2AFI
 
 | | Crystal Structure of MgADP bound Av2-Av1 Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2005-07-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein
Science, 309, 2005
|
|
5CFK
 
 | | Crystal structure of Proliferating Cell Nuclear Antigen from Leishmania donovani at 3.2 A resolution | | Descriptor: | Proliferating cell nuclear antigen,Proliferating cell nuclear antigen | | Authors: | Shukla, P.K, Yadav, S.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-07-08 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Proliferating Cell Nuclear Antigen from Leishmania donovani at 3.2 A resolution
To Be Published
|
|
3G71
 
 | | Co-crystal structure of Bruceantin bound to the large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10E, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
1XPE
 
 | | HIV-1 subtype B genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
3G4S
 
 | | Co-crystal structure of Tiamulin bound to the large ribosomal subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L10e, ... | | Authors: | Gurel, G, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome.
J.Mol.Biol., 389, 2009
|
|
1XTI
 
 | | Structure of Wildtype human UAP56 | | Descriptor: | ISOPROPYL ALCOHOL, Probable ATP-dependent RNA helicase p47 | | Authors: | Shi, H, Cordin, O, Minder, C.M, Linder, P, Xu, R.M. | | Deposit date: | 2004-10-21 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the human ATP-dependent splicing and export factor UAP56
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
2Y7P
 
 | | DntR Inducer Binding Domain in Complex with Salicylate. Trigonal crystal form | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-HYDROXYBENZOIC ACID, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Devesse, L, Smirnova, I, Lonneborg, R, Kapp, U, Brzezinski, P, Leonard, G.A, Dian, C. | | Deposit date: | 2011-02-01 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Dntr Inducer Binding Domains in Complex with Salicylate Offer Insights Into the Activation of Lysr-Type Transcriptional Regulators.
Mol.Microbiol., 81, 2011
|
|
1ZL3
 
 | | Coupling of active site motions and RNA binding | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*CP*GP*GP*UP*(FLO) UP*CP*GP*AP*UP*CP*CP*CP*GP*UP*UP*GP*C)-3', SULFATE ION, tRNA pseudouridine synthase B | | Authors: | Hoang, C, Hamilton, C.S, Mueller, E.G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain
Protein Sci., 14, 2005
|
|
6EXN
 
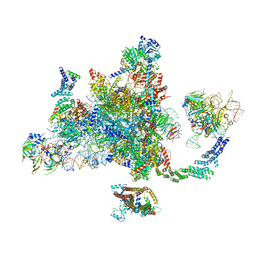 | | Post-catalytic P complex spliceosome with 3' splice site docked | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
1ZDY
 
 | |
3B7G
 
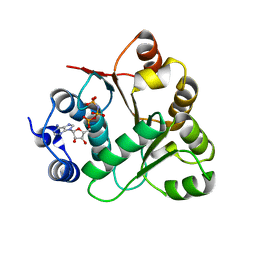 | | Human DEAD-box RNA helicase DDX20, Conserved domain I (DEAD) in complex with AMPPNP (Adenosine-(Beta,gamma)-imidotriphosphate) | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP-dependent RNA helicase DDX20 | | Authors: | Karlberg, T, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
1ZFX
 
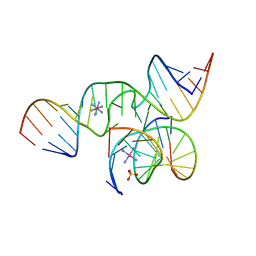 | | The Structure of a minimal all-RNA Hairpin Ribozyme with the mutant G8U at the cleavage site | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*UP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Wedekind, J.E. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Water in the Active Site of an All-RNA Hairpin Ribozyme and Effects of Gua8 Base Variants on the Geometry of Phosphoryl Transfer.
Biochemistry, 45, 2006
|
|
1ZJB
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 (monoclinic form) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Trehalulose synthase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2005-04-28 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, purification, crystallization and preliminary X-ray crystallographic studies of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3B5R
 
 | |
2AFH
 
 | | Crystal Structure of Nucleotide-Free Av2-Av1 Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2005-07-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein
Science, 309, 2005
|
|
3BIM
 
 | |
5CQG
 
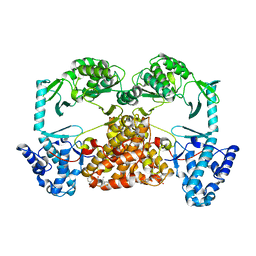 | | Structure of Tribolium telomerase in complex with the highly specific inhibitor BIBR1532 | | Descriptor: | 2-{[(2E)-3-(naphthalen-2-yl)but-2-enoyl]amino}benzoic acid, Telomerase reverse transcriptase | | Authors: | Bryan, C, Rice, C, Hoffman, H, Harkisheimer, M, Sweeney, M, Skordalakes, E. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Telomerase Inhibition by the Highly Specific BIBR1532.
Structure, 23, 2015
|
|
3B66
 
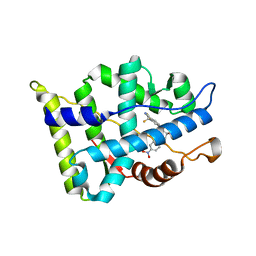 | | Crystal structure of the androgen receptor ligand binding domain in complex with SARM S-21 | | Descriptor: | 4-{[(1R,2S)-1,2-dihydroxy-2-methyl-3-(4-nitrophenoxy)propyl]amino}-2-(trifluoromethyl)benzonitrile, Androgen receptor | | Authors: | Bohl, C.E, Miller, D.D, Dalton, J.T. | | Deposit date: | 2007-10-27 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Effect of B-ring substitution pattern on binding mode of propionamide selective androgen receptor modulators
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1XTK
 
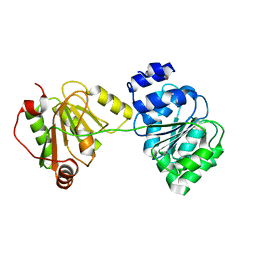 | | structure of DECD to DEAD mutation of human UAP56 | | Descriptor: | BETA-MERCAPTOETHANOL, Probable ATP-dependent RNA helicase p47 | | Authors: | Shi, H, Cordin, O, Minder, C.M, Linder, P, Xu, R.M. | | Deposit date: | 2004-10-22 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human ATP-dependent splicing and export factor UAP56
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XP7
 
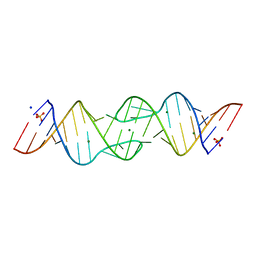 | | HIV-1 subtype F genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1XPF
 
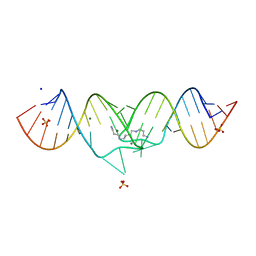 | | HIV-1 subtype A genomic RNA Dimerization Initiation Site | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ennifar, E, Dumas, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Polymorphism of Bulged-out Residues in HIV-1 RNA DIS Kissing Complex and Structure Comparison with Solution Studies
J.Mol.Biol., 356, 2006
|
|
1XRH
 
 | |
1XSZ
 
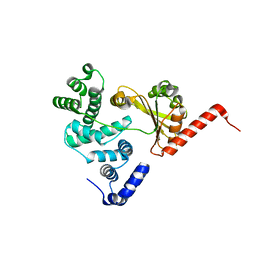 | | The structure of RalF | | Descriptor: | guanine nucleotide exchange protein | | Authors: | Amor, J.C, Swails, J, Roy, C.R, Nagai, H, Ingmundson, A, Cheng, X, Kahn, R.A. | | Deposit date: | 2004-10-20 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site
J.Biol.Chem., 280, 2005
|
|
3B45
 
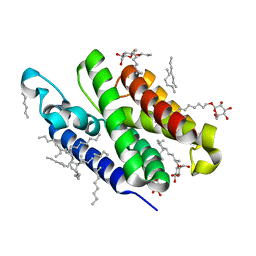 | | Crystal structure of GlpG at 1.9A resolution | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
3B6Y
 
 | | Crystal Structure of the Second HIN-200 Domain of Interferon-Inducible Protein 16 | | Descriptor: | Gamma-interferon-inducible protein Ifi-16, SULFATE ION | | Authors: | Liao, J.C.C, Lam, R, Ravichandran, M, Duan, S, Tempel, W, Chirgadze, N.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure Analysis of the Second HIN Domain of IFI16.
To be Published
|
|
