5D8B
 
 | |
6EQP
 
 | | Human butyrylcholinesterase in complex with ethopropazine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nachon, F, Brazzolotto, X, Wandhammer, M, Trovaslet-Leroy, M, Rosenberry, T.L, Macdonald, I.R, Darvesh, S. | | Deposit date: | 2017-10-14 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.349417 Å) | | Cite: | Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study.
Molecules, 22, 2017
|
|
6EP4
 
 | | Human butyrylcholinesterase in complex with decamethonium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nachon, F, Brazzolotto, X, Wandhammer, M, Trovaslet-Leroy, M, Rosenberry, T.L, Macdonald, I.R, Darvesh, S. | | Deposit date: | 2017-10-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.30005383 Å) | | Cite: | Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study.
Molecules, 22, 2017
|
|
5DUV
 
 | | Crystal structure of the human galectin-4 N-terminal carbohydrate recognition domain in complex with lactose | | Descriptor: | ACETATE ION, CALCIUM ION, Galectin-4, ... | | Authors: | Bum-Erdene, K, Blanchard, H. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterisation of human galectin-4 N-terminal carbohydrate recognition domain in complex with glycerol, lactose, 3'-sulfo-lactose, and 2'-fucosyllactose.
Sci Rep, 6, 2016
|
|
1JOD
 
 | |
5DUW
 
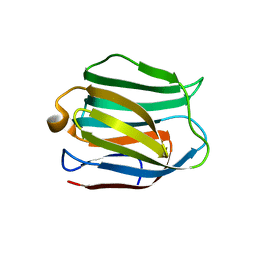 | |
4KXP
 
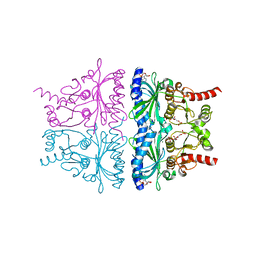 | | Crystal Structure of AMP complexes of Porcine Liver Fructose-1,6-bisphosphatase Mutant I10D in T-state | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Iancu, C.V, Mukund, S, Choe, J.-Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2013-05-27 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Displacement of a Catalytically Essential Loop from the Active Site of Mammalian Fructose-1,6-bisphosphatase.
Biochemistry, 52, 2013
|
|
5DUX
 
 | | Crystal structure of the human galectin-4 N-terminal carbohydrate recognition domain in complex with 2'-fucosyllactose | | Descriptor: | FORMIC ACID, GLYCEROL, Galectin-4, ... | | Authors: | Bum-Erdene, K, Blanchard, H. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterisation of human galectin-4 N-terminal carbohydrate recognition domain in complex with glycerol, lactose, 3'-sulfo-lactose, and 2'-fucosyllactose.
Sci Rep, 6, 2016
|
|
5DOX
 
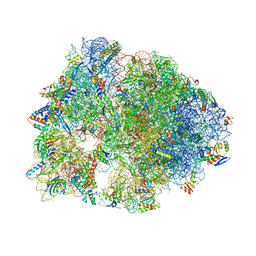 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
5E7K
 
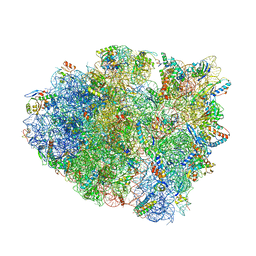 | | Structure of T. thermophilus 70S ribosome complex with mRNA and cognate tRNALys in the A-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-10-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
3LK8
 
 | |
1ZDU
 
 | | The Crystal Structure of Human Liver Receptor Homologue-1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2, ... | | Authors: | Wang, W, Zhang, C, Marimuthu, A, Krupka, H.I, Tabrizizad, M, Shelloe, R, Mehra, U, Eng, K, Nguyen, H, Settachatgul, C, Powell, B, Milburn, M.V, West, B.L. | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ZDT
 
 | | The Crystal Structure of Human Steroidogenic Factor-1 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, Steroidogenic factor 1 | | Authors: | Wang, W, Zhang, C, Marimuthu, A, Krupka, H.I, Tabrizizad, M, Shelloe, R, Mehra, U, Eng, K, Nguyen, H, Settachatgul, C, Powell, B, Milburn, M.V, West, B.L. | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5ZV5
 
 | | P domain of GII.17-2014/15 complexed with A-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
8HJ1
 
 | | GPR21(wt) and Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
4AXN
 
 | | Hallmarks of processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases | | Descriptor: | ACETATE ION, CALCIUM ION, CHITINASE C1 | | Authors: | Payne, C.M, Baban, J, Synstad, B, Backe, P.H, Arvai, A.S, Dalhus, B, Bjoras, M, Eijsink, V.G.H, Sorlie, M, Beckham, G.T, Vaaje-Kolstad, G. | | Deposit date: | 2012-06-13 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Hallmarks of Processivity in Glycoside Hydrolases from Crystallographic and Computational Studies of the Serratia Marcescens Chitinases.
J.Biol.Chem., 287, 2012
|
|
4BDB
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
8SDJ
 
 | |
4BFL
 
 | |
1CCL
 
 | |
4K7N
 
 | | HUMAN PEROXIREDOXIN 5 with a fragment | | Descriptor: | 4-METHYLCATECHOL, Peroxiredoxin-5, mitochondrial | | Authors: | Guichou, J.F. | | Deposit date: | 2013-04-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Plos One, 9, 2014
|
|
4AYM
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 3 P106A mutant | | Descriptor: | COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
4K0A
 
 | |
1BJ9
 
 | | EFFECT OF UNNATURAL HEME SUBSTITUTION ON KINETICS OF ELECTRON TRANSFER IN CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, [7,12-DEACETYL-3,8,13,17-TETRAMETHYL-21H,23H-PORPHINE-2,18-DIPROPANOATO(2-)-N21,N22,N23,N24]-IRON | | Authors: | Miller, M.A, Kraut, J. | | Deposit date: | 1998-07-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of Unnatural Heme Substitution on Kinetics of Electron Transfer in Cytochrome C Peroxidase
To be Published
|
|
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
