2YLX
 
 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: Asp66Ala MUTANT IN COMPLEX WITH NADP AND MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
2YNX
 
 | | Crystal Structure of Ancestral Thioredoxin Relative to Last Archaea Common Ancestor (LACA) from the Precambrian Period | | Descriptor: | ACETATE ION, LACA THIOREDOXIN, SODIUM ION | | Authors: | Gavira, J.A, Ingles-Prieto, A, Ibarra-Molero, B, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-10-19 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Conservation of Protein Structure Over Four Billion Years
Structure, 21, 2013
|
|
7LB6
 
 | | PDX1.2/PDX1.3 co-expression complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2 | | Authors: | Novikova, I.V, Evans, J.E. | | Deposit date: | 2021-01-07 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Tunable Heteroassembly of a Plant Pseudoenzyme-Enzyme Complex.
Acs Chem.Biol., 16, 2021
|
|
7DYT
 
 | | Human JMJD5 in complex with MN and 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid. | | Descriptor: | 5-((4-methoxybenzyl)amino)pyridine-2,4-dicarboxylic acid, Bifunctional peptidase and arginyl-hydroxylase JMJD5, MANGANESE (II) ION | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2021-01-23 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 5-Substituted Pyridine-2,4-dicarboxylate Derivatives Have Potential for Selective Inhibition of Human Jumonji-C Domain-Containing Protein 5.
J.Med.Chem., 66, 2023
|
|
5MIJ
 
 | | X-ray structure of carboplatin-encapsulated horse spleen apoferritin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Pontillo, N, Ferraro, G, Helliwell, J.R, Merlino, A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | X-ray Structure of the Carboplatin-Loaded Apo-Ferritin Nanocage.
ACS Med Chem Lett, 8, 2017
|
|
2FSU
 
 | | Crystal Structure of the PhnH Protein from Escherichia Coli | | Descriptor: | ACETATE ION, Protein phnH, SODIUM ION | | Authors: | Adams, M.A, Luo, Y, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-23 | | Release date: | 2007-03-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnH: an essential component of carbon-phosphorus lyase in Escherichia coli.
J.Bacteriol., 190, 2008
|
|
7LZ7
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5k | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
2PKR
 
 | | Crystal structure of (A+CTE)4 chimeric form of photosyntetic glyceraldehyde-3-phosphate dehydrogenase, complexed with NADP | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase Aor, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Fermani, S, Falini, G, Ripamonti, A. | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7LZ8
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5t | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)pyrido[3,2-d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
7LM6
 
 | | Crystal structure of the Zn(II)-bound AdcAII H205L mutant variant of Streptococcus pneumoniae | | Descriptor: | Adhesion protein, ZINC ION | | Authors: | Luo, Z, Zupan, M, McDevitt, C.A, Kobe, B. | | Deposit date: | 2021-02-05 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.367 Å) | | Cite: | Conformation of the Solute-Binding Protein AdcAII Influences Zinc Uptake in Streptococcus pneumoniae .
Front Cell Infect Microbiol, 11, 2021
|
|
2FU8
 
 | | Zinc-beta-lactamase L1 from stenotrophomonas maltophilia (d-captopril complex) | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, Metallo-beta-lactamase L1, ... | | Authors: | Nauton, L, Garau, G, Kahn, R, Dideberg, O. | | Deposit date: | 2006-01-26 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the design of inhibitors for the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia.
J.Mol.Biol., 375, 2008
|
|
7LM5
 
 | | Crystal structure of the Zn(II)-bound AdcAII H65A mutant variant of Streptococcus pneumoniae | | Descriptor: | Adhesion protein, CHLORIDE ION, SODIUM ION, ... | | Authors: | Luo, Z, Zupan, M, McDevitt, C.A, Kobe, B. | | Deposit date: | 2021-02-05 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformation of the Solute-Binding Protein AdcAII Influences Zinc Uptake in Streptococcus pneumoniae .
Front Cell Infect Microbiol, 11, 2021
|
|
7GE1
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-6af13d92-3 (Mpro-x11294) | | Descriptor: | 3C-like proteinase, 5-methoxy-N-[2-(2-methoxyphenoxy)ethyl]-2-oxo-1,2-dihydroquinoline-4-carboxamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2PHM
 
 | | STRUCTURE OF PHENYLALANINE HYDROXYLASE DEPHOSPHORYLATED | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE-4-HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
3I1U
 
 | | Carboxypeptidase A Inhibited by a Thiirane Mechanism-Based inactivator | | Descriptor: | (2S,3R)-2-benzyl-3-sulfanylbutanoic acid, Carboxypeptidase A1 (Pancreatic), GLYCEROL, ... | | Authors: | Fernandez, D. | | Deposit date: | 2009-06-28 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | The X-ray structure of carboxypeptidase A inhibited by a thiirane mechanism-based inhibitor
Chem.Biol.Drug Des., 75, 2010
|
|
5MNQ
 
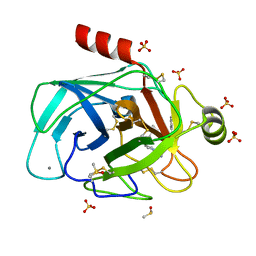 | | Cationic trypsin in complex with a derivative of N-amidinopiperidine | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(1-carbamimidoylpiperidin-4-yl)methyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.337 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
7LM7
 
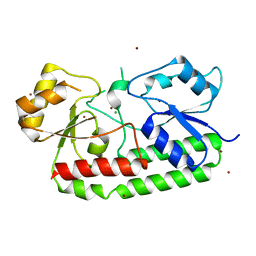 | | Crystal structure of the Zn(II)-bound AdcAII E280Q mutant variant of Streptococcus pneumoniae | | Descriptor: | Adhesion protein, CHLORIDE ION, ZINC ION | | Authors: | Luo, Z, Zupan, M, McDevitt, C.A, Kobe, B. | | Deposit date: | 2021-02-05 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Conformation of the Solute-Binding Protein AdcAII Influences Zinc Uptake in Streptococcus pneumoniae .
Front Cell Infect Microbiol, 11, 2021
|
|
3I28
 
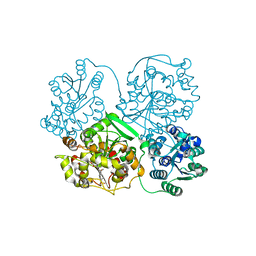 | | Crystal Structure of soluble epoxide Hydrolase | | Descriptor: | 4-cyano-N-{(3S)-3-(4-fluorophenyl)-3-[4-(methylsulfonyl)phenyl]propyl}benzamide, Epoxide hydrolase 2 | | Authors: | Farrow, N.A. | | Deposit date: | 2009-06-29 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based optimization of arylamides as inhibitors of soluble epoxide hydrolase.
J.Med.Chem., 52, 2009
|
|
5MO6
 
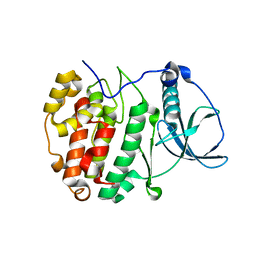 | | Crystal Structure of CK2alpha with N-(3-(((2-chloro-[1,1'-biphenyl]-4-yl)methyl)amino)propyl)methanesulfonamide bound | | Descriptor: | 3-[3-[(3-chloranyl-4-phenyl-phenyl)methylamino]propylamino]-3-oxidanylidene-propanoic acid, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
2YVX
 
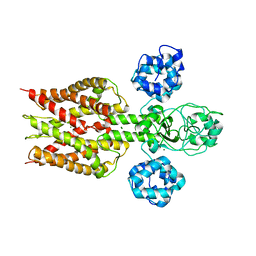 | | Crystal structure of magnesium transporter MgtE | | Descriptor: | MAGNESIUM ION, Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
5MSG
 
 | |
3HZX
 
 | |
2PQF
 
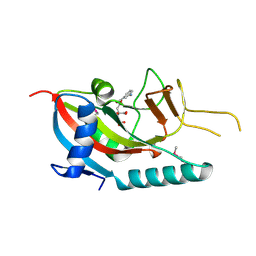 | | Human Poly(ADP-Ribose) Polymerase 12, Catalytic fragment in complex with an inhibitor 3-Aminobenzoic acid | | Descriptor: | 3-AMINOBENZOIC ACID, CITRIC ACID, Poly [ADP-ribose] polymerase 12 | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hogbom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-02 | | Release date: | 2007-05-15 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
2PR9
 
 | | Mu2 adaptin subunit (AP50) of AP2 adaptor (second domain), complexed with GABAA receptor-gamma2 subunit-derived internalization peptide DEEYGYECL | | Descriptor: | AP-2 complex subunit mu-1, GABA(A) receptor subunit gamma-2 peptide | | Authors: | Vahedi-Faridi, A, Haucke, V, Kittler, J.T, Kukhtina, V, Moss, S.J, Saenger, W, Chen, G.-J, Tretter, V, Smith, K, Yan, Z, McAinsh, K, Arancibia-Carcamo, L. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor gamma2 subunit.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3I09
 
 | |
