4L1Q
 
 | |
1W1B
 
 | |
2L2Y
 
 | | Thiostrepton, epimer form of residue 9 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2I0R
 
 | |
4NMK
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Boyko, K.M, Bezsudnova, E.Y, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. crystallized in microgravity (complex with NADP+)
To be Published
|
|
2F10
 
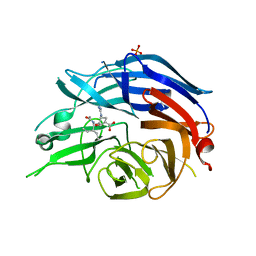 | | Crystal Structure of the Human Sialidase Neu2 in Complex with Peramivir inhibitor | | Descriptor: | 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, McKimm-Breschkin, J, Colman, P.M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with Peramivir inhibitor
To be Published
|
|
2F1O
 
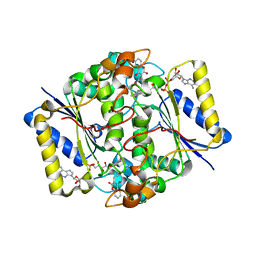 | | Crystal Structure of NQO1 with Dicoumarol | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Shaul, Y, Asher, G, Dym, O, Tsvetkov, P, Adler, J, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-11-15 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol.
Biochemistry, 45, 2006
|
|
4NQ3
 
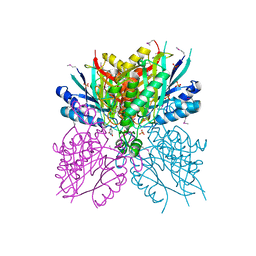 | | Crystal structure of cyanuic acid hydrolase from A. caulinodans | | Descriptor: | BARBITURIC ACID, Cyanuric acid amidohydrolase, MAGNESIUM ION, ... | | Authors: | Cho, S, Shi, K, Aihara, H. | | Deposit date: | 2013-11-23 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Cyanuric acid hydrolase from Azorhizobium caulinodans ORS 571: crystal structure and insights into a new class of Ser-Lys dyad proteins.
Plos One, 9, 2014
|
|
4NY2
 
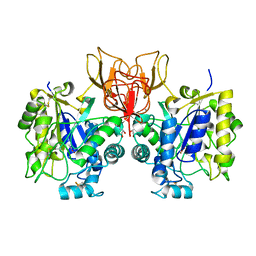 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2JSY
 
 | | Solution structure of Tpx in the oxidized state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2F5N
 
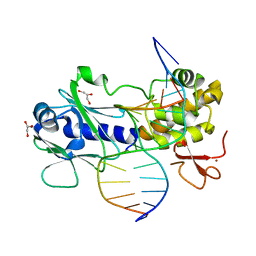 | | MutM crosslinked to undamaged DNA sampling A:T base pair IC1 | | Descriptor: | 5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', GLYCEROL, ... | | Authors: | Banerjee, A, Santos, W.L, Verdine, G.L. | | Deposit date: | 2005-11-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a DNA glycosylase searching for lesions.
Science, 311, 2006
|
|
2K2P
 
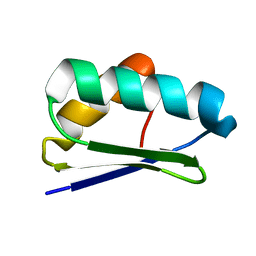 | | Solution NMR structure of protein Atu1203 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium (NESG) target AtT10, Ontario Center for Structural Proteomics target ATC1183 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Lemak, A, Gutmanas, A, Yee, A, Semesi, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein Atu1203 from Agrobacterium tumefaciens.
To be Published
|
|
4NDH
 
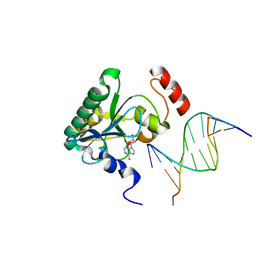 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4M81
 
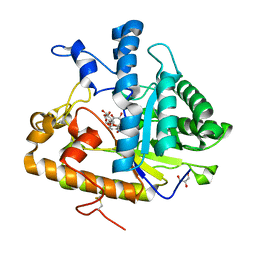 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution | | Descriptor: | 4-nitrophenyl beta-D-glucopyranoside, EXO-1,3-BETA-GLUCANASE, GLYCEROL, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
2F7O
 
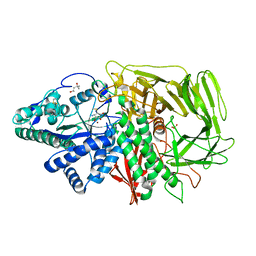 | | Golgi alpha-mannosidase II complex with mannostatin A | | Descriptor: | (1R,2R,3R,4S,5R)-4-AMINO-5-(METHYLTHIO)CYCLOPENTANE-1,2,3-TRIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-07-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Basis of the Inhibition of Golgi alpha-Mannosidase II by Mannostatin A and the Role of the Thiomethyl Moiety in Ligand-Protein Interactions.
J.Am.Chem.Soc., 128, 2006
|
|
1W9A
 
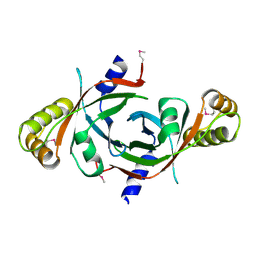 | | Crystal structure of Rv1155 from Mycobacterium tuberculosis | | Descriptor: | PUTATIVE PYRIDOXINE/PYRIDOXAMINE 5'-PHOSPHATE OXIDASE | | Authors: | Cannan, S, Sulzenbacher, G, Roig-Zamboni, V, Scappuccini, L, Frassinetti, F, Maurien, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Rv1155 from Mycobacterium Tuberculosis
FEBS Lett., 579, 2005
|
|
4M8U
 
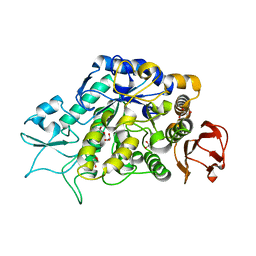 | | The Structure of MalL mutant enzyme V200A from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-13 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
4NFV
 
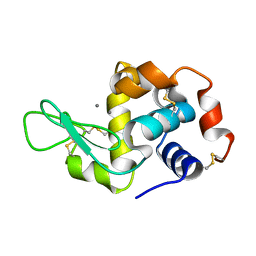 | | Previously de-ionized HEW lysozyme batch crystallized in 1.1 M MnCl2 | | Descriptor: | CHLORIDE ION, Lysozyme C, MANGANESE (II) ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3FWG
 
 | | Ferric camphor bound Cytochrome P450cam, Arg365Leu, Glu366Gln, monoclinic crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Schlichting, I, Von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2FFZ
 
 | |
4NKV
 
 | |
4MDR
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain D190A mutant, in complex with a sorting peptide from the amyloid precursor protein (APP) | | Descriptor: | AP-4 complex subunit mu-1, Amyloid beta A4 protein | | Authors: | Ross, B.H, Lin, Y, Corales, E.A, Burgos, P.V, Mardones, G.A. | | Deposit date: | 2013-08-23 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Characterization of Cargo-Binding Sites on the mu 4-Subunit of Adaptor Protein Complex 4.
Plos One, 9, 2014
|
|
4NMM
 
 | | Crystal Structure of a G12C Oncogenic Variant of Human KRas Bound to a Novel GDP Competitive Covalent Inhibitor | | Descriptor: | 5'-O-[(S)-{[(S)-[2-(acetylamino)ethoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]guanosine, GTPase KRas, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Lim, S.M, Westover, K.D. | | Deposit date: | 2013-11-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2FH8
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with isomaltose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
1WVA
 
 | | Crystal structure of human arginase I from twinned crystal | | Descriptor: | Arginase 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Di Costanzo, L, Sabio, G, Mora, A, Rodriguez, P.C, Ochoa, A.C, Centeno, F, Christianson, D.W. | | Deposit date: | 2004-12-14 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of human arginase I at 1.29 A resolution and exploration of inhibition in the immune response
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
