4KRT
 
 | |
2I0K
 
 | |
4KTF
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with 4,4'-(3-amino-1H-pyrazole-4,5-diyl)diphenol | | Descriptor: | 4,4'-(3-amino-1H-pyrazole-4,5-diyl)diphenol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A. | | Deposit date: | 2013-05-20 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Overcoming the Limitations of Fragment Merging: Rescuing a Strained Merged Fragment Series Targeting Mycobacterium tuberculosis CYP121.
Chemmedchem, 8, 2013
|
|
4DUB
 
 | | cytochrome P450 BM3h-9D7 MRI sensor bound to dopamine | | Descriptor: | L-DOPAMINE, PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 9D7 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
3E84
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
4NC5
 
 | | Human sialidase 2 in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, PHOSPHATE ION, Sialidase-2 | | Authors: | Buchini, S, Gallat, F.-X, Greig, I.R, Kim, J.-H, Wakatsuki, S, Chavas, L.M.G, Withers, S.G. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.513 Å) | | Cite: | Tuning mechanism-based inactivators of neuraminidases: mechanistic and structural insights.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2I3A
 
 | | Crystal structure of N-Acetyl-gamma-Glutamyl-Phosphate Reductase (Rv1652) from Mycobacterium tuberculosis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Moraidin, F, James, M.N.G, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of N-acetyl-gamma-glutamyl-phosphate Reductase from Mycobacterium tuberculosis in Complex with NADP(+).
J.Mol.Biol., 367, 2007
|
|
4KVL
 
 | | Crystal structure of Oryza sativa fatty acid alpha-dioxygenase Y379F with palmitic acid | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fatty acid alpha-oxidase, ... | | Authors: | Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of alpha-dioxygenase from Oryza sativa: Insights into substrate binding and activation by hydrogen peroxide.
Protein Sci., 22, 2013
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
4NOV
 
 | | Xsa43E, a GH43 family enzyme from Butyrivibrio proteoclasticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Xylosidase/arabinofuranosidase Xsa43E | | Authors: | Till, M, Arcus, V.L. | | Deposit date: | 2013-11-20 | | Release date: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural analysis of the GH43 enzyme Xsa43E from Butyrivibrio proteoclasticus
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
4NDI
 
 | | Human Aprataxin (Aptx) AOA1 variant K197Q bound to RNA-DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3', 5'-R(P*G)-D(P*TP*TP*AP*TP*GP*AP*TP*TP*C)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4KTZ
 
 | | Lactococcus phage 67 RuvC | | Descriptor: | MAGNESIUM ION, RuvC endonuclease | | Authors: | Rafferty, J.B, Green, V. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutants of phage bIL67 RuvC with enhanced Holliday junction binding selectivity and resolution symmetry.
Mol.Microbiol., 89, 2013
|
|
6ASK
 
 | |
3EBA
 
 | | CAbHul6 FGLW mutant (humanized) in complex with human lysozyme | | Descriptor: | CAbHul6, Lysozyme C, SULFATE ION | | Authors: | Loris, R, Vincke, C, Saerens, D, Martinez-Rodriguez, S, Muyldermans, S, Conrath, K. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | General Strategy to Humanize a Camelid Single-domain Antibody and Identification of a Universal Humanized Nanobody Scaffold
J.Biol.Chem., 284, 2009
|
|
1W36
 
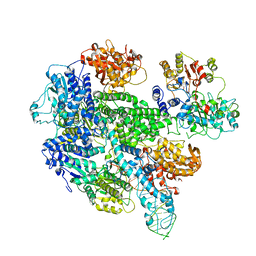 | | RecBCD:DNA complex | | Descriptor: | CALCIUM ION, DNA HAIRPIN, EXODEOXYRIBONUCLEASE V ALPHA CHAIN, ... | | Authors: | Singleton, M.R, Dillingham, M.S, Gaudier, M, C Kowalczykowski, S, Wigley, D.B. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Recbcd Enzyme Reveals a Machine for Processing DNA Breaks
Nature, 432, 2004
|
|
4NGJ
 
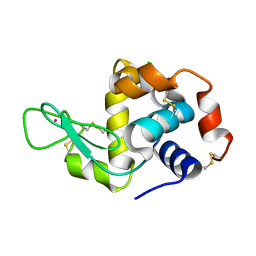 | | Dialyzed HEW lysozyme batch crystallized in 1.0 M RbCl and collected at 100 K | | Descriptor: | CHLORIDE ION, Lysozyme C, RUBIDIUM ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-02 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4KZO
 
 | |
4KWL
 
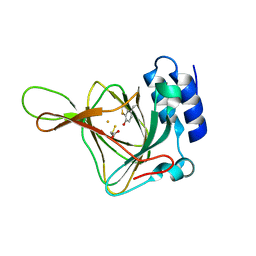 | | Rat cysteine dioxygenase with 3-mercaptopropionic acid persulfide bound | | Descriptor: | 3-disulfanylpropanoic acid, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Souness, R.J, Wilbanks, S.M, Jameson, G.B, Jameson, G.N.L. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanistic implications of persulfenate and persulfide binding in the active site of cysteine dioxygenase.
Biochemistry, 52, 2013
|
|
6AWA
 
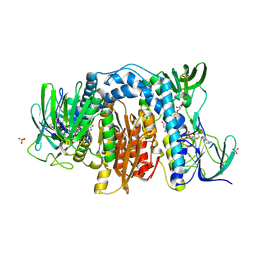 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
2I82
 
 | |
2HXC
 
 | |
4KX6
 
 | | Plasticity of the quinone-binding site of the complex II homolog quinol:fumarate reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Singh, P.K, Sarwar, M, Maklashina, E, Kotlyar, V, Rajagukguk, S, Tomasiak, T.M, Cecchini, G, Iverson, T.M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasticity of the Quinone-binding Site of the Complex II Homolog Quinol:Fumarate Reductase.
J.Biol.Chem., 288, 2013
|
|
1W61
 
 | |
6AX6
 
 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, IODIDE ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
4NJM
 
 | |
