2OIY
 
 | |
3CN6
 
 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S274E mutant | | Descriptor: | Aquaporin, CADMIUM ION | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Tornroth-Horsefield, S. | | Deposit date: | 2008-03-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
7G00
 
 | | Crystal Structure of human FABP1 in complex with 2-[[3-(5-tert-butyl-1,2,4-oxadiazol-3-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]cyclopentene-1-carboxylic acid | | Descriptor: | 2-{[(3P)-3-(5-tert-butyl-1,2,4-oxadiazol-3-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, liver, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Neidhart, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
2B1V
 
 | | Human estrogen receptor alpha ligand-binding domain in complex with OBCP-1M and a glucocorticoid receptor interacting protein 1 NR box II peptide | | Descriptor: | 4-[(1S,2S,5S)-5-(HYDROXYMETHYL)-8-METHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2005-09-16 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
4BKS
 
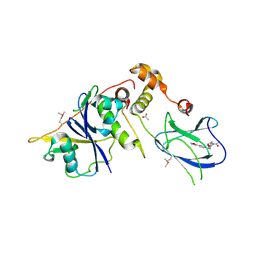 | | von Hippel Lindau protein:ElonginB:ElonginC complex, in complex with (2S,4R)-1-ethanoyl-N-[[4-(1,3-oxazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide | | Descriptor: | (2S,4R)-1-ethanoyl-N-[[4-(1,3-oxazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, ACETATE ION, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Van Molle, I, Dias, D.M, Baud, M, Galdeano, C, Geraldes, C.F.G.C, Ciulli, A. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Is NMR Fragment Screening Fine-Tuned to Assess Druggability of Protein-Protein Interactions?
Acs Med.Chem.Lett., 5, 2014
|
|
5AWD
 
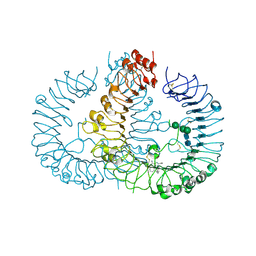 | | Crystal structure of human TLR8 in complex with N1-4-aminomethylbenzyl (IMDQ) | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Design of Human TLR8-Specific Agonists with Augmented Potency and Adjuvanticity.
J.Med.Chem., 58, 2015
|
|
5RET
 
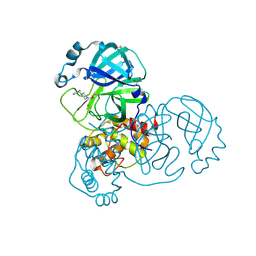 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102269 | | Descriptor: | 1-{4-[(3-chlorophenyl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6C0M
 
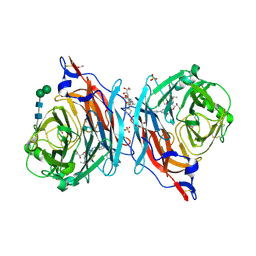 | | The synthesis, biological evaluation and structural insights of unsaturated 3-N-substituted sialic acids as probes of human parainfluenza virus-3 haemagglutinin-neuraminidase | | Descriptor: | 1,2-ETHANEDIOL, 2,6-anhydro-3,5-dideoxy-5-[(2-methylpropanoyl)amino]-3-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-D-galacto-non-2-enoni c acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, Ve, T, von Itzstein, M. | | Deposit date: | 2018-01-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insights into Human Parainfluenza Virus 3 Hemagglutinin-Neuraminidase Using Unsaturated 3- N-Substituted Sialic Acids as Probes.
ACS Chem. Biol., 13, 2018
|
|
5R8Q
 
 | |
4FE6
 
 | | Crystal Structure of HIV-1 Protease in Complex with an enamino-oxindole inhibitor | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[({(3Z)-3-[1-(methylamino)ethylidene]-2-oxo-2,3-dihydro-1H-indol-5-yl}sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, HIV protease | | Authors: | Silva, A.M, Eissenstat, M, Guerassina, T, Gulnik, S, Afonina, E, Yu, B, Erickson, J, Ludke, D, Yokoe, H. | | Deposit date: | 2012-05-29 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enamino-oxindole HIV protease inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5RWW
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2241115980 | | Descriptor: | 1-[(furan-2-yl)methyl]-4-(methylsulfonyl)piperazine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
3USR
 
 | | Structure of Y194F glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Issoglio, F.M, Carrizo, M.E, Romero, J.M, Curtino, J.A. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of monomeric and dimeric glycogenin autoglucosylation.
J.Biol.Chem., 287, 2012
|
|
3FZN
 
 | | Intermediate analogue in benzoylformate decarboxylase | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, CHLORIDE ION, ... | | Authors: | Bruning, M, Berheide, M, Meyer, D, Golbik, R, Bartunik, H, Liese, A, Tittmann, K. | | Deposit date: | 2009-01-26 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural and kinetic studies on native intermediates and an intermediate analogue in benzoylformate decarboxylase reveal a least motion mechanism with an unprecedented short-lived predecarboxylation intermediate.
Biochemistry, 48, 2009
|
|
3UWC
 
 | | Structure of an aminotransferase (DegT-DnrJ-EryC1-StrS family) from Coxiella burnetii in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-12-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4HZG
 
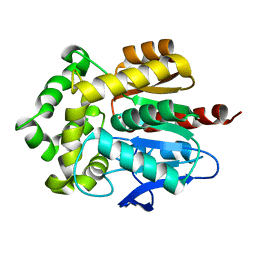 | | Structure of haloalkane dehalogenase DhaA from Rhodococcus rhodochrous | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Stsiapanava, A, Weiss, M.S, Mesters, J.R, Kuta Smatanova, I. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2CQD
 
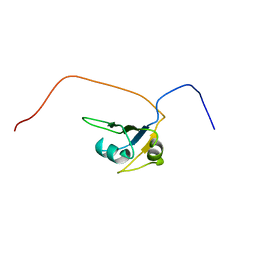 | | Solution Structure of the RNA recognition motif in RNA-binding region containing protein 1 | | Descriptor: | RNA-binding region containing protein 1 | | Authors: | Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RNA recognition motif in RNA-binding region containing protein 1
To be Published
|
|
7FUR
 
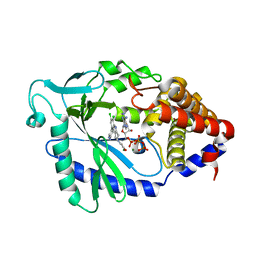 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydropyrido[4,3-b]indol-2-yl]-2-hydroxyethanone | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
20GS
 
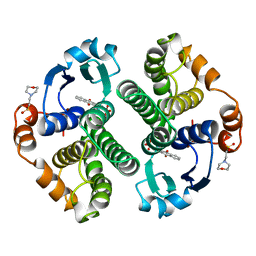 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
3G7E
 
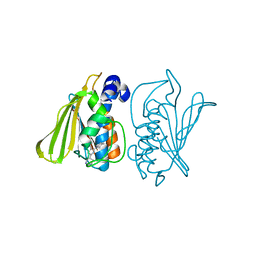 | |
1G5A
 
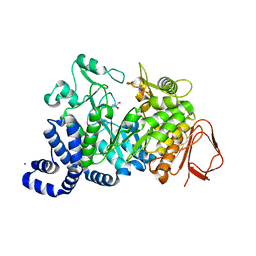 | | AMYLOSUCRASE FROM NEISSERIA POLYSACCHAREA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMYLOSUCRASE, ... | | Authors: | Skov, L.K, Mirza, O, Henriksen, A, De Montalk, G.P, Remaud-Simeon, M, Sarcabal, P, Willemot, R.-M, Monsan, P, Gajhede, M. | | Deposit date: | 2000-10-31 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Amylosucrase, A Glucan-synthesizing Enzyme from the alpha-Amylase Family
J.Biol.Chem., 276, 2001
|
|
1G8O
 
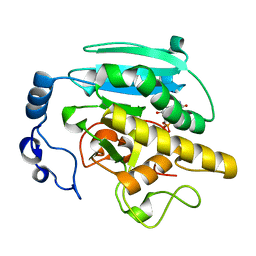 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NATIVE BOVINE ALPHA-1,3-GALACTOSYLTRANSFERASE CATALYTIC DOMAIN | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Gastinel, L.N, Bigon, C, Misra, A.K, Hindsgaul, O, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-11-20 | | Release date: | 2001-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
3GPJ
 
 | | Crystal structure of the yeast 20S proteasome in complex with syringolin B | | Descriptor: | N-{[(1S)-2-methyl-1-{[(5S,8S)-5-(1-methylethyl)-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]carbamoyl}propyl]carbamoyl}-L-valine, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Huber, R, Kaiser, M. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2FTR
 
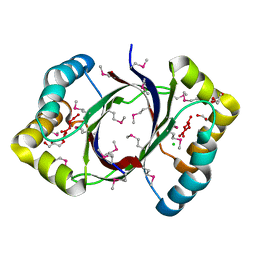 | |
5RFH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102277 | | Descriptor: | 1-{4-[(5-chlorothiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
2D19
 
 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the kissing-loop dimer | | Descriptor: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
