6XMP
 
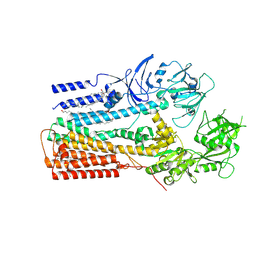 | | Structure of P5A-ATPase Spf1, Apo form | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, P5A-type ATPase | | Authors: | Park, E, Sim, S.I. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase.
Science, 369, 2020
|
|
6WVI
 
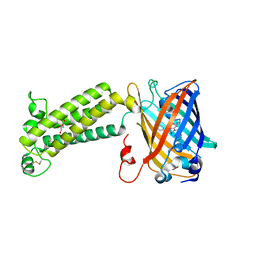 | | VKOR-like from Takifugu rubripes | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVH
 
 | | Human VKOR with Brodifacoum | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Brodifacoum, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVG
 
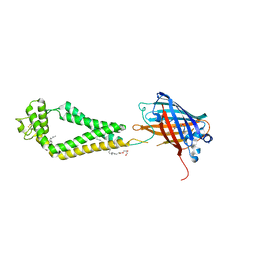 | | human CD53 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein, ... | | Authors: | Yang, Y, Liu, S, Li, W. | | Deposit date: | 2020-05-06 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Open conformation of tetraspanins shapes interaction partner networks on cell membranes.
Embo J., 39, 2020
|
|
6WVF
 
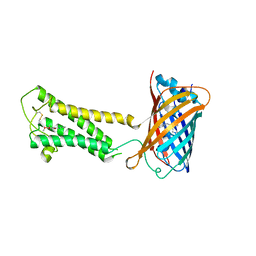 | | E.coli DsbB C104S with ubiquinone | | Descriptor: | Green fluorescent protein,Disulfide bond formation protein B,Green fluorescent protein, UBIQUINONE-1 | | Authors: | Liu, S, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Termini restraining of small membrane proteins enables structure determination at near-atomic resolution.
Sci Adv, 6, 2020
|
|
6WVE
 
 | | Chicken SPCS1 | | Descriptor: | Green fluorescent protein,Chicken Signal Peptidase Complex Subunit 1,Green fluorescent protein | | Authors: | Liu, S, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Termini restraining of small membrane proteins enables structure determination at near-atomic resolution.
Sci Adv, 6, 2020
|
|
6WVD
 
 | | Human JAGN1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Green fluorescent protein, Protein jagunal homolog 1 chimera | | Authors: | Yang, Y, Liu, S, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Termini restraining of small membrane proteins enables structure determination at near-atomic resolution.
Sci Adv, 6, 2020
|
|
6WVB
 
 | | Takifugu rubripes VKOR-like with warfarin | | Descriptor: | S-WARFARIN, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVA
 
 | |
6WV9
 
 | |
6WV8
 
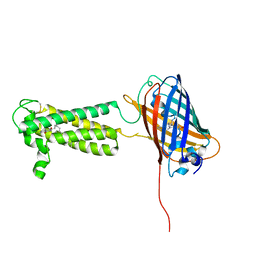 | | Takifugu rubripes VKOR-like C138S mutant with vitamin K1 | | Descriptor: | PHYLLOQUINONE, Vitamin K epoxide reductase-like protein, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV7
 
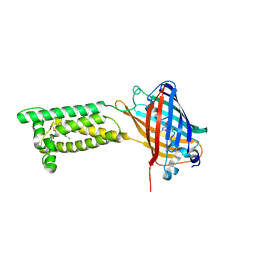 | | Human VKOR with Chlorophacinone | | Descriptor: | Chlorophacinone, Vitamin K epoxide reductase, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV6
 
 | | Human VKOR with phenindione | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phenindione, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV5
 
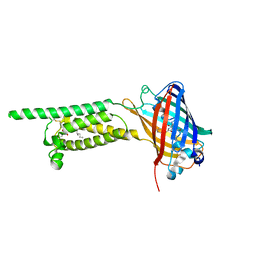 | | Human VKOR C43S mutant with vitamin K1 epoxide | | Descriptor: | (2R,3R)-2-hydroxy-3-methyl-2-[(2E,7S)-3,7,11,15-tetramethylhexadec-2-en-1-yl]-2,3-dihydronaphthalene-1,4-dione, Vitamin K epoxide reductase Cys43Ser mutant, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV4
 
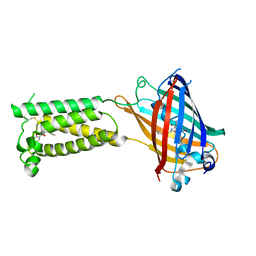 | | Human VKOR C43S with warfarin | | Descriptor: | S-WARFARIN, Vitamin K epoxide reductase Cys43Ser mutant, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.012 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WV3
 
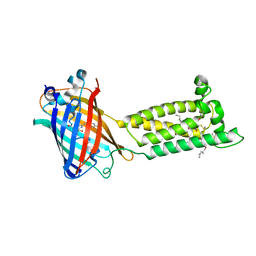 | | Human VKOR with warfarin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, S-WARFARIN, Vitamin K epoxide reductase, ... | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WSG
 
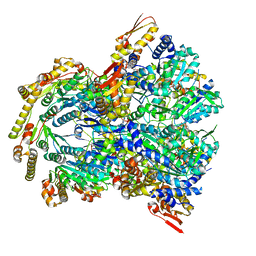 | | ClpX-ClpP complex bound to ssrA-tagged GFP, intermediate complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, Green fluorescent protein, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
6WRF
 
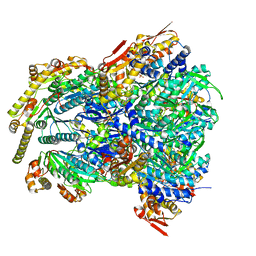 | | ClpX-ClpP complex bound to GFP-ssrA, recognition complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
6WR2
 
 | |
6WNK
 
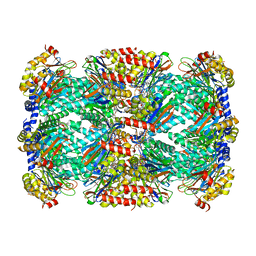 | | Macrocyclic peptides TDI5575 that selectively inhibit the Mycobacterium tuberculosis proteasome | | Descriptor: | (12S,15S)-N-[(2-fluorophenyl)methyl]-10,13-dioxo-12-{2-oxo-2-[(2R)-2-phenylpyrrolidin-1-yl]ethyl}-2-oxa-11,14-diazatricyclo[15.2.2.1~3,7~]docosa-1(19),3(22),4,6,17,20-hexaene-15-carboxamide, CITRIC ACID, DIMETHYLFORMAMIDE, ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Macrocyclic Peptides that Selectively Inhibit the Mycobacterium tuberculosis Proteasome.
J.Med.Chem., 64, 2021
|
|
6W24
 
 | | ClpA Engaged2 State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W23
 
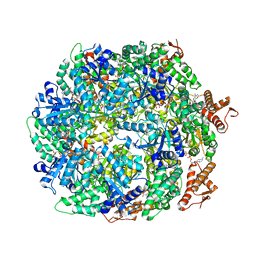 | | ClpA Disengaged State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W22
 
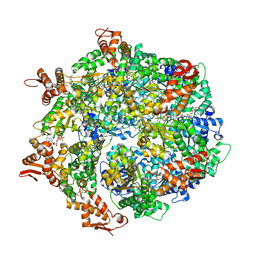 | | ClpA Engaged1 State bound to RepA-GFP (ClpA Focused Refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W21
 
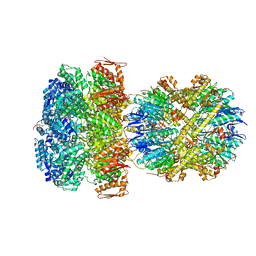 | | ClpAP Engaged2 State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W20
 
 | | ClpAP Disengaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
