1GSE
 
 | |
2V0I
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
4DA6
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with ganciclovir | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
3MQH
 
 | | crystal structure of the 3-N-acetyl transferase WlbB from Bordetella petrii in complex with CoA and UDP-3-amino-2-acetamido-2,3-dideoxy glucuronic acid | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-4-amino-6-{[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxytetrahydro-2H-pyran-2-carboxylic acid, 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | thoden, J.B, holden, H.M. | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular structure of WlbB, a bacterial N-acetyltransferase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid .
Biochemistry, 49, 2010
|
|
7H6H
 
 | | THE 1.94 A CRYSTAL STRUCTURE OF HUMAN CATHEPSIN G IN COMPLEX WITH 1-[(7-fluoronaphthalen-1-yl)methyl]-3-[[methoxycarbonyl(methyl)amino]methyl]indole-2-carboxylic acid | | Descriptor: | 1-[(7-fluoronaphthalen-1-yl)methyl]-3-{[(methoxycarbonyl)(methyl)amino]methyl}-1H-indole-2-carboxylic acid, Cathepsin G, SULFATE ION, ... | | Authors: | Banner, D.W, Benz, J.M, Schlatter, D, Hilpert, H. | | Deposit date: | 2024-04-19 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of human Chymase and Cathepsin G
To be published
|
|
3A3H
 
 | | CELLOTRIOSE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHERANS AT 1.6 A RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Davies, G.J, Brzozowski, A.M, Andersen, K, Schulein, M. | | Deposit date: | 1998-02-01 | | Release date: | 1999-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
4KE6
 
 | | Crystal structure D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in complex with 1-rac-lauroyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
3Q2I
 
 | | Crystal structure of the WlbA dehydrognase from Chromobactrium violaceum in complex with NADH and UDP-GlcNAcA at 1.50 A resolution | | Descriptor: | (2S,3S,4R,5R,6R)-5-acetamido-6-[[[(2R,3S,4R,5R)-5-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-hydroxy-phosphoryl]oxy-3,4-dihydroxy-oxane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and Structural Characterization of WlbA from Bordetella pertussis and Chromobacterium violaceum: Enzymes Required for the Biosynthesis of 2,3-Diacetamido-2,3-dideoxy-d-mannuronic Acid.
Biochemistry, 50, 2011
|
|
4HAY
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K548E,K579Q)-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Exportin-1, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HC6
 
 | | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, H.-B, Han, G.-W, Hung, L.-W, Terwilliger, C.T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mycobacterium tuberculosis Rv2523cE77A x-ray structure solved with 1.8 angstrom resolution
To be Published
|
|
4HCF
 
 | | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Copper Bound from Bacillus anthracis | | Descriptor: | COPPER (II) ION, Cupredoxin 1, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-29 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Copper Bound from Bacillus anthracis
To be Published
|
|
3VVH
 
 | |
5QK5
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1267773786 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
3PO0
 
 | | Crystal structure of SAMP1 from Haloferax volcanii | | Descriptor: | ACETATE ION, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Jeong, Y.J, Jeong, B.-C, Song, H.K. | | Deposit date: | 2010-11-21 | | Release date: | 2011-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
1WEM
 
 | | Solution structure of PHD domain in death inducer-obliterator 1(DIO-1) | | Descriptor: | Death associated transcription factor 1, ZINC ION | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PHD domain in death inducer-obliterator 1(DIO-1)
To be Published
|
|
1X63
 
 | |
5E97
 
 | | Glycoside Hydrolase ligand structure 1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5QP1
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1190363272 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3M7W
 
 | | Crystal Structure of Type I 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 in Covalent Complex with Dehydroquinate | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
4FPD
 
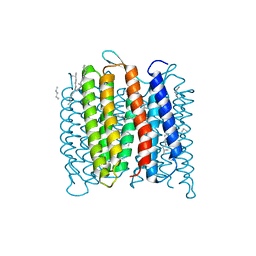 | | Deprotonation of D96 in bacteriorhodopsin opens the proton uptake pathway | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, CHLORIDE ION, ... | | Authors: | Wang, T, Sessions, A.O, Lunde, C.S, Rouani, S, Glaeser, R.M, Facciotti, M.T, Duan, Y. | | Deposit date: | 2012-06-22 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Deprotonation of d96 in bacteriorhodopsin opens the proton uptake pathway.
Structure, 21, 2013
|
|
3A32
 
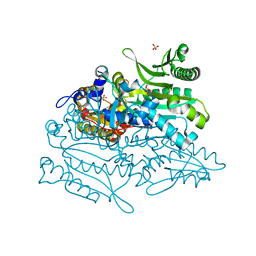 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix | | Descriptor: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | Authors: | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2009-06-07 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
4K6T
 
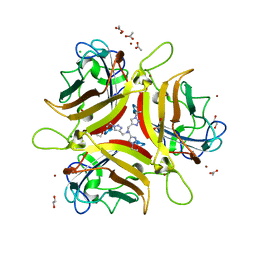 | | Crystal structure of Ad37 fiber knob in complex with trivalent sialic acid inhibitor ME0385 | | Descriptor: | 1,2-ETHANEDIOL, 2,2',2''-[nitrilotris(methanediyl-1H-1,2,3-triazole-4,1-diyl)]triethanol, ACETATE ION, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2013-04-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
2WC3
 
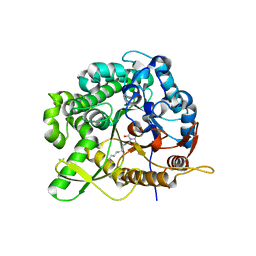 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-8-epi-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8S,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
3COP
 
 | |
1LBV
 
 | | Crystal Structure of apo-form (P21) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | Descriptor: | fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
