4WSL
 
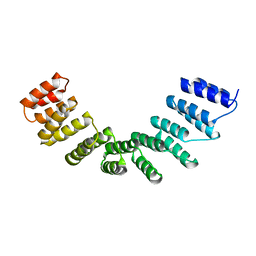 | | Crystal structure of designed cPPR-polyC protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4Y7T
 
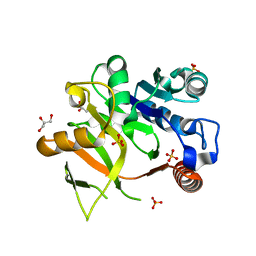 | | Structural analysis of MurU | | Descriptor: | GLYCEROL, Nucleotidyl transferase, SULFATE ION | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4Y7U
 
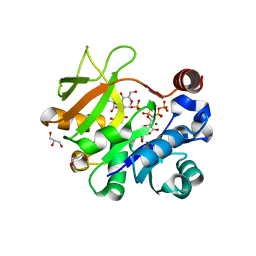 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, GLYCEROL, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4Y7V
 
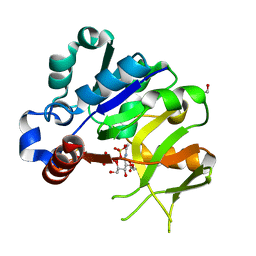 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, GLYCEROL, IMIDODIPHOSPHORIC ACID, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4YDW
 
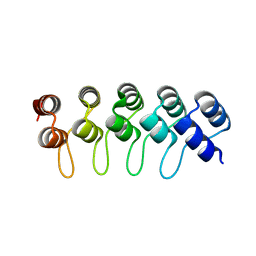 | |
5VNW
 
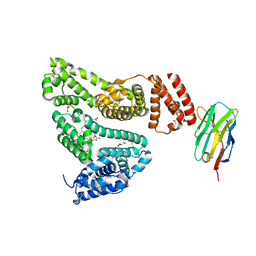 | |
5VNV
 
 | | Crystal structure of Nb.b201 | | Descriptor: | FORMIC ACID, Nb.b201, PENTAETHYLENE GLYCOL, ... | | Authors: | Kruse, A.C, McMahon, C. | | Deposit date: | 2017-05-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Yeast surface display platform for rapid discovery of conformationally selective nanobodies.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WOV
 
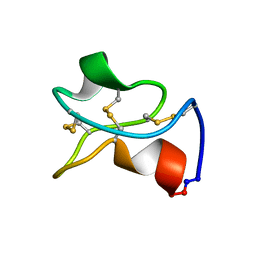 | |
5WRX
 
 | | VG13P structure in LPS | | Descriptor: | analogue peptide VG13P | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2016-12-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Insights into a Glycine-Mediated Short Analogue of a Designed Peptide in Lipopolysaccharide Micelles: Correlation Between Compact Structure and Anti-Endotoxin Activity.
Biochemistry, 56, 2017
|
|
5WOW
 
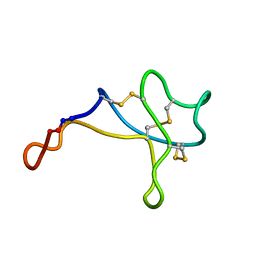 | |
5XG5
 
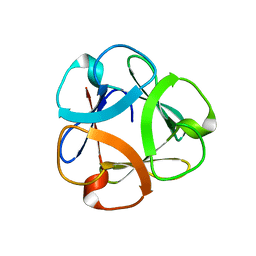 | | Crystal structure of Mitsuba-1 with bound NAcGal | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MITSUBA-1 | | Authors: | Terada, D, Voet, A.R.D, Kamata, K, Zhang, K.Y.J, Tame, J.R.H. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Computational design of a symmetrical beta-trefoil lectin with cancer cell binding activity.
Sci Rep, 7, 2017
|
|
5XNG
 
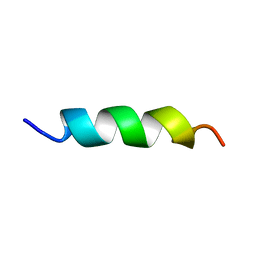 | | EFK17A structure in Microgel MAA60 | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2017-05-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Aspects of High Content Packing of Antimicrobial Peptides in Polymer Microgels
ACS Appl Mater Interfaces, 9, 2017
|
|
6ANM
 
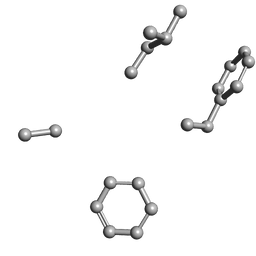 | |
5W4H
 
 | |
5W4I
 
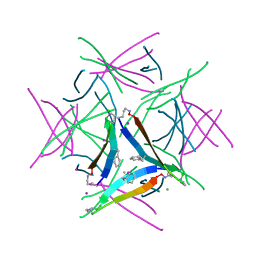 | |
5W4J
 
 | |
6ANN
 
 | |
5US3
 
 | |
5VAV
 
 | |
5VL4
 
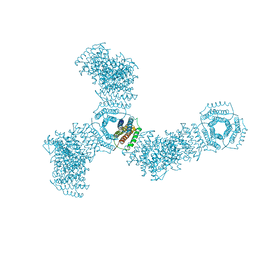 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
5YSC
 
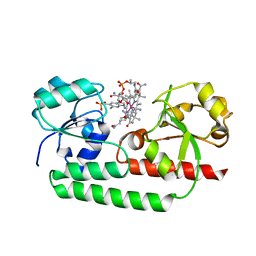 | |
8VE7
 
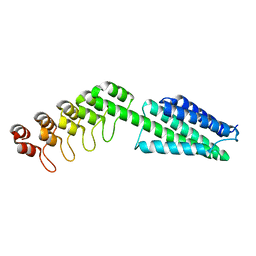 | |
8VDZ
 
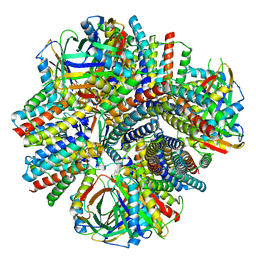 | | A designed tetrahedral protein scaffold - DARP14 | | Descriptor: | Subunit A, Subunit B | | Authors: | Suder, D.S, Gonen, S. | | Deposit date: | 2023-12-18 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mitigating the Blurring Effect of CryoEM Averaging on a Flexible and Highly Symmetric Protein Complex through Sub-Particle Reconstruction.
Int J Mol Sci, 25, 2024
|
|
9AZI
 
 | |
9ATC
 
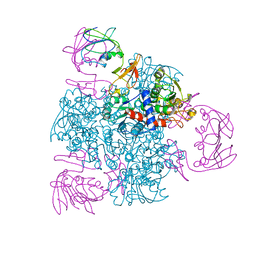 | | ATCASE Y165F MUTANT | | Descriptor: | ASPARTATE TRANSCARBAMOYLASE, ZINC ION | | Authors: | Ha, Y, Allewell, N.M. | | Deposit date: | 1998-06-26 | | Release date: | 1999-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Intersubunit hydrogen bond acts as a global molecular switch in Escherichia coli aspartate transcarbamoylase.
Proteins, 33, 1998
|
|
