8APG
 
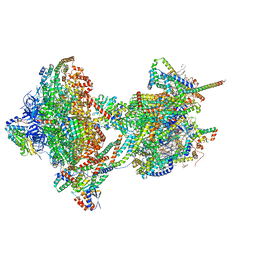 | | rotational state 2b of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
2JD7
 
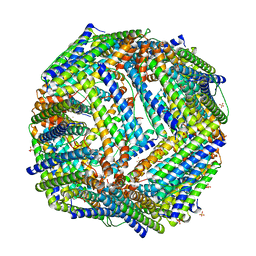 | |
8APN
 
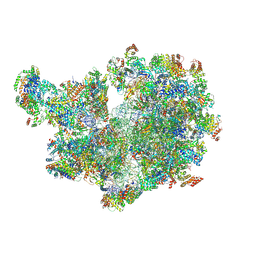 | | Structure of the mitochondrial ribosome from Polytomella magna with tRNA bound to the P site | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hypothetical protein, MAGNESIUM ION, ... | | Authors: | Tobiasson, V, Berzina, I, Amunts, A. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a mitochondrial ribosome with fragmented rRNA in complex with membrane-targeting elements.
Nat Commun, 13, 2022
|
|
7CKB
 
 | | Simplified Alpha-Carboxysome, T=3 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7CKC
 
 | | Simplified Alpha-Carboxysome, T=4 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
2JKR
 
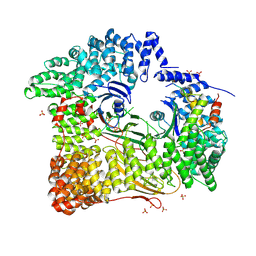 | | AP2 CLATHRIN ADAPTOR CORE with Dileucine peptide RM(phosphoS)QIKRLLSE | | Descriptor: | AP-2 COMPLEX SUBUNIT ALPHA-2, AP-2 COMPLEX SUBUNIT BETA-1, AP-2 COMPLEX SUBUNIT MU-1, ... | | Authors: | Owen, D.J, McCoy, A.J, Kelly, B.T, Evans, P.R. | | Deposit date: | 2008-08-29 | | Release date: | 2008-10-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A Structural Explanation for the Binding of Endocytic Dileucine Motifs by the Ap2 Complex.
Nature, 456, 2008
|
|
8ADL
 
 | | Cryo-EM structure of the SEA complex | | Descriptor: | Maintenance of telomere capping protein 5, Nitrogen permease regulator 2, Nitrogen permease regulator 3, ... | | Authors: | Tafur, L, Loewith, R. | | Deposit date: | 2022-07-08 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of the SEA complex.
Nature, 611, 2022
|
|
5L5H
 
 | |
6Y9D
 
 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate L-Carnitine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARNITINE, Carnitine monooxygenase oxygenase subunit, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
5L5O
 
 | | Yeast 20S proteasome with human beta5i (1-138) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
6Y3D
 
 | | X-ray structure of thermophilic C-phycocyanin from Galdiera phlegrea | | Descriptor: | ACETATE ION, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Ferraro, G, Lucignano, R, Marseglia, A, Merlino, A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of C-phycocyanin from Galdieria phlegrea: Determinants of thermostability and comparison with a C-phycocyanin in the entire phycobilisome.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
5L65
 
 | |
5L5V
 
 | |
6UXE
 
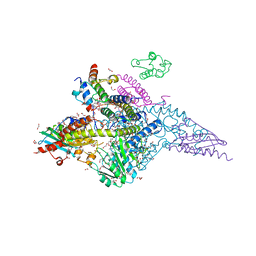 | | Structure of the human mitochondrial desulfurase complex Nfs1-ISCU2(M140I)-ISD11 with E.coli ACP1 at 1.57 A resolution showing flexibility of N terminal end of ISCU2 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2019-11-07 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
5L6B
 
 | | Yeast 20S proteasome with mouse beta5i (1-138) and mouse beta6 (97-111; 118-133) in complex with ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L8B
 
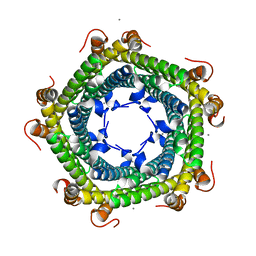 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
6V96
 
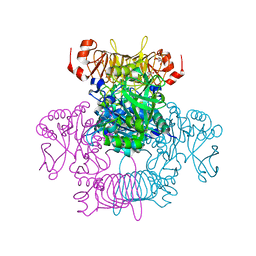 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72E | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Hussien, R, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6Y69
 
 | |
6YCX
 
 | | Plasmodium falciparum Myosin A full-length, pre-powerstroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin A tail domain interacting protein, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Krementsova, E.B, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | Full-length Plasmodium falciparum myosin A and essential light chain PfELC structures provide new anti-malarial targets.
Elife, 9, 2020
|
|
6YFD
 
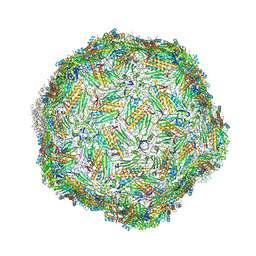 | | Virus-like particle of Beihai levi-like virus 14 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YBS
 
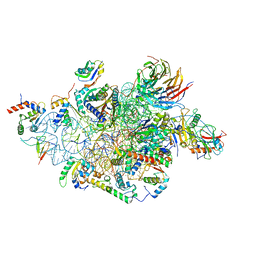 | | Structure of a human 48S translational initiation complex - head | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
8ALY
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament (G1032W mutant) | | Descriptor: | Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Mariotti, L, Inian, O, Desfosses, A, Beuron, F, Morris, E.P, Guettler, S. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of tankyrase activation by polymerization.
Nature, 612, 2022
|
|
5KPW
 
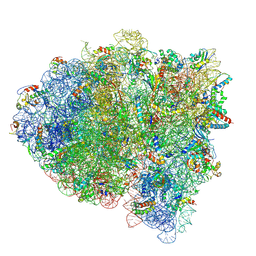 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
6VCS
 
 | | SRA domain of UHRF1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*AP*CP*AP*GP*GP*C)-3'), E3 ubiquitin-protein ligase UHRF1, UNK-UNK-UNK-UNK, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-22 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SRA domain of UHRF1 in complex with DNA
To Be Published
|
|
8AS0
 
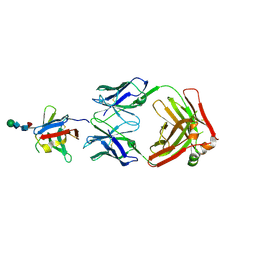 | | PD-1 extracellular domain in complex with Fab fragment from D12 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ongaro, T, Scietti, L, Pluss, L, Peissert, F, Villa, A, Puca, E, De Luca, R, Neri, D, Forneris, F. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Selection of a PD-1 blocking antibody from a novel fully human phage display library.
Protein Sci., 31, 2022
|
|
