7NGL
 
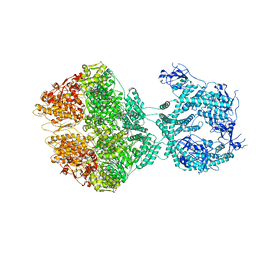 | | R-state of wild type human mitochondrial LONP1 protease bound to endogenous ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
1DE7
 
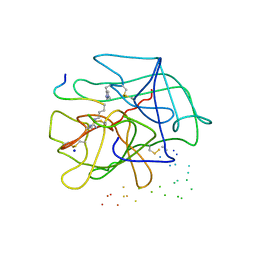 | |
3L4H
 
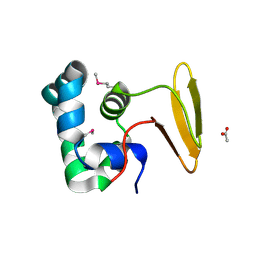 | | Helical box domain and second WW domain of the human E3 ubiquitin-protein ligase HECW1 | | Descriptor: | ACETIC ACID, E3 ubiquitin-protein ligase HECW1 | | Authors: | Walker, J.R, Qiu, L, Li, Y, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The tandem helical box and second WW domains of human HECW1
To be Published
|
|
4GPG
 
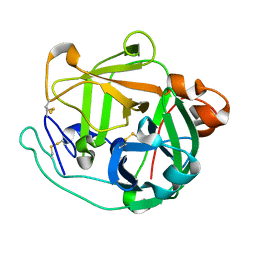 | | X/N joint refinement of Achromobacter Lyticus Protease I free form at pD8.0 | | Descriptor: | Protease 1 | | Authors: | Ohnishi, Y, Yamada, T, Kurihara, K, Tanaka, I, Sakiyama, F, Masaki, T, Niimura, N. | | Deposit date: | 2012-08-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | NEUTRON DIFFRACTION (1.895 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of Achromobacter protease I at pD 8.0: protonation states and hydration structure in the free-form.
Biochim.Biophys.Acta, 1834, 2013
|
|
4HSS
 
 | |
1ZJK
 
 | | Crystal structure of the zymogen catalytic region of human MASP-2 | | Descriptor: | Mannan-binding lectin serine protease 2 | | Authors: | Gal, P, Harmat, V, Kocsis, A, Bian, T, Barna, L, Ambrus, G, Vegh, B, Balczer, J, Sim, R.B, Naray-Szabo, G, Zavodszky, P. | | Deposit date: | 2005-04-29 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A True Autoactivating Enzyme: Structural insight into mannose-binding lectin-associated serine protease-2 activations
J.Biol.Chem., 280, 2005
|
|
2B4I
 
 | |
2B5T
 
 | | 2.1 Angstrom structure of a nonproductive complex between antithrombin, synthetic heparin mimetic SR123781 and two S195A thrombin molecules | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Johnson, D.J, Li, W, Luis, S.A, Carrell, R.W, Huntington, J.A. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation.
J.Biol.Chem., 281, 2006
|
|
2AIQ
 
 | |
2B4H
 
 | |
6U7Q
 
 | | NMR solution structure of SFTI-R10 | | Descriptor: | GLY-ARG-CYS-THR-LYS-SER-ILE-PRO-PRO-ARG-CYS-PHE-PRO-ASP inhibitor | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U24
 
 | | NMR solution structure of triazole bridged SFTI-1 | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-07-01 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7U
 
 | | NMR solution structure of triazole bridged matriptase inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ARG-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U7S
 
 | |
6U2J
 
 | | EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
1QQ4
 
 | |
6U7W
 
 | | NMR solution structure of a triazole bridged KLK7 inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-LYS-ALA-LEU-PHE-SER-ASN-PRO-PRO-ILE-ALA-PHE-PRO-ASN | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U2W
 
 | | EM structure of MPEG-1(L425K) pre-pore complex bound to liposome | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
1QGF
 
 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3R, 4S)N-PARA-TOLUENESULPHONYL-3-ETHYL-4-(CARBOXYLIC ACID)PYRROLIDIN-2-ONE | | Descriptor: | (2S,3S)-3-FORMYL-2-({[(4-METHYLPHENYL)SULFONYL]AMINO}METHYL)PENTANOIC ACID, CALCIUM ION, ELASTASE, ... | | Authors: | Wilmouth, R.C, Kassamally, S, Westwood, N.J, Sheppard, R.J, Claridge, T.D.W, Wright, P.A, Pritchard, G.J, Schofield, C.J. | | Deposit date: | 1999-04-27 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic insights into the inhibition of serine proteases by monocyclic lactams.
Biochemistry, 38, 1999
|
|
6UFN
 
 | |
8GO6
 
 | | Fungal immunomodulatory protein FIP-nha N39A | | Descriptor: | fungal immunomodulatory protein FIP-nha N39A | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
8GO7
 
 | | Fungal immunomodulatory protein FIP-nha N5+39A | | Descriptor: | Fungal immunomodulatory protein FIP-nha | | Authors: | Liu, Y, Bastiaan-Net, S, Hoppenbrouwers, T, Li, Z, Wichers, H.J. | | Deposit date: | 2022-08-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Glycosylation Contributes to Thermostability and Proteolytic Resistance of rFIP-nha ( Nectria haematococca ).
Molecules, 28, 2023
|
|
6UHV
 
 | |
6UII
 
 | |
8GO5
 
 | |
