7XNR
 
 | | SARS-CoV-2 Omicron BA.2.13 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
3BSG
 
 | |
2X5X
 
 | | The crystal structure of PhaZ7 at atomic (1.2 Angstrom) resolution reveals details of the active site and suggests a substrate binding mode | | Descriptor: | CHLORIDE ION, IODIDE ION, PHB DEPOLYMERASE PHAZ7, ... | | Authors: | Wakadkar, S, Hermawan, S, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of PhaZ7 at atomic (1.2 A) resolution reveals details of the active site and suggests a substrate-binding mode.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 66, 2010
|
|
3L4Z
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with Salacinol | | Descriptor: | 1,4-DIDEOXY-1,4-[[2R,3R)-2,4-DIHYDROXY-3-(SULFOXY)BUTYL]EPISULFONIUMYLIDENE]-D-ARABINITOL INNER SALT, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New glucosidase inhibitors from an ayurvedic herbal treatment for type 2 diabetes: structures and inhibition of human intestinal maltase-glucoamylase with compounds from Salacia reticulata.
Biochemistry, 49, 2010
|
|
3BL1
 
 | | Carbonic anhydrase inhibitors. Sulfonamide diuretics revisited old leads for new applications | | Descriptor: | 4-chloro-N-[(2S)-2-methyl-2,3-dihydro-1H-indol-1-yl]-3-sulfamoylbenzamide, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Temperini, C, Cecchi, A, Supuran, C.T. | | Deposit date: | 2007-12-10 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbonic anhydrase inhibitors. Interaction of indapamide and related diuretics with 12 mammalian isozymes and X-ray crystallographic studies for the indapamide-isozyme II adduct.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3Q9X
 
 | | Crystal structure of human CK2 alpha in complex with emodin at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, ... | | Authors: | Battistutta, R, Ranchio, A, Papinutto, E. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the flexible regions of the catalytic alpha-subunit of protein kinase CK2
To be Published
|
|
1V9M
 
 | | Crystal structure of the C subunit of V-type ATPase from Thermus thermophilus | | Descriptor: | GLYCEROL, V-type ATP synthase subunit C | | Authors: | Numoto, N, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the C subunit of V-type ATPase from Thermus thermophilus at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6N9R
 
 | | Structure of the Quorum Quenching lactonase from Parageobacillus caldoxylosilyticus bound to substrate 3-oxo-C12-AHL | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Elias, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structural Determinants Accounting for the Broad Substrate Specificity of the Quorum Quenching Lactonase GcL.
Chembiochem, 20, 2019
|
|
5SM7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1247413608 | | Descriptor: | 1-(methanesulfonyl)piperidin-4-ol, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6N9I
 
 | | Structure of the Quorum Quenching lactonase from Parageobacillus caldoxylosilyticus - free | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Bergonzi, C, Schwab, M, Elias, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structural Determinants Accounting for the Broad Substrate Specificity of the Quorum Quenching Lactonase GcL.
Chembiochem, 20, 2019
|
|
7AW4
 
 | | MerTK kinase domain with type 3 inhibitor from a DNA-encoded library | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-(4-methyl-1,2,3-thiadiazol-5-yl)-N-((R)-1-(((R)-3-(methylamino)-3-oxo-1-(4-(trifluoromethyl)phenyl)propyl)amino)-1-oxo-4-phenylbutan-2-yl)isoxazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
2QE7
 
 | | Crystal structure of the f1-atpase from the thermoalkaliphilic bacterium bacillus sp. ta2.a1 | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit epsilon, ... | | Authors: | Stocker, A, Keis, S, Vonck, J, Cook, G.M, Dimroth, P. | | Deposit date: | 2007-06-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Structural Basis for Unidirectional Rotation of Thermoalkaliphilic F(1)-ATPase.
Structure, 15, 2007
|
|
5L5Q
 
 | |
7DZN
 
 | |
2VB8
 
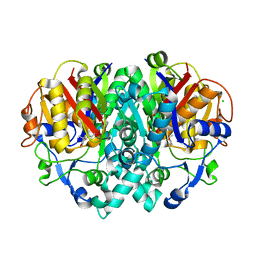 | | beta-ketoacyl-ACP synthase I (KAS) from E. coli with bound inhibitor thiolactomycin | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, CHLORIDE ION, THIOLACTOMYCIN | | Authors: | Pappenberger, G, Schulz-Gasch, T, Bailly, J, Hennig, M. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-Assisted Discovery of an Aminothiazole Derivative as a Lead Molecule for Inhibition of Bacterial Fatty-Acid Synthesis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5SLC
 
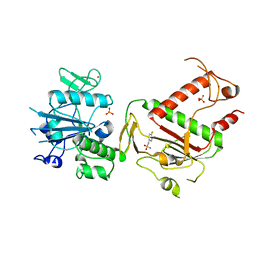 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1849009686 | | Descriptor: | 1-(4-fluoro-2-methylphenyl)methanesulfonamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLO
 
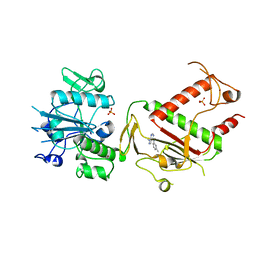 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z56983806 | | Descriptor: | 1-methyl-N-(3-methylphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3MAC
 
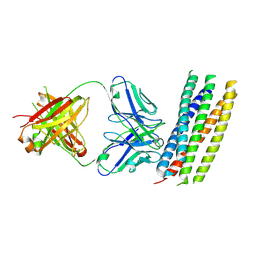 | | crystal structure of GP41-derived protein complexed with fab 8062 | | Descriptor: | Fab8062, Transmembrane glycoprotein | | Authors: | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
7B8E
 
 | | Torpedo californica acetylcholinesterase complexed with Ca+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L. | | Deposit date: | 2020-12-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
4NL1
 
 | |
5ZGF
 
 | | Crystal structure of NDM-1 Q123G mutant | | Descriptor: | HYDROXIDE ION, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
7DZM
 
 | |
5SMH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434938 | | Descriptor: | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4NHC
 
 | |
4I41
 
 | | Crystal Structure of human Ser/Thr kinase Pim1 in complex with mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Xie, Y, Huang, N. | | Deposit date: | 2012-11-27 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new target for an old drug: identifying mitoxantrone as a nanomolar inhibitor of PIM1 kinase via kinome-wide selectivity modeling.
J. Med. Chem., 56, 2013
|
|
